We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox2QRU
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 7: | Line 7: | ||
2QRU is known to be a hydrolase, most active on esterase. The structure is 276 amino acids long, which is approximately 30.14 kDa. Knowing 2QRU is an alpha/beta hydrolase, the literature helped give further information on possible experimental conditions and substrates that can be used to identify activity. However, experimental data has not been obtained yet. | 2QRU is known to be a hydrolase, most active on esterase. The structure is 276 amino acids long, which is approximately 30.14 kDa. Knowing 2QRU is an alpha/beta hydrolase, the literature helped give further information on possible experimental conditions and substrates that can be used to identify activity. However, experimental data has not been obtained yet. | ||
| + | |||
== Structure/Sequence Analysis == | == Structure/Sequence Analysis == | ||
'''Sequence Analysis''' | '''Sequence Analysis''' | ||
| + | |||
'''SPRITE''' | '''SPRITE''' | ||
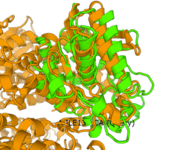
| - | [[Image:2QRU(green)2LPR(yellow).png | thumb]]Analysis with Sprite displayed the proteins closest in structure to 2QRU. To do this, Sprite analyzed 2QRU for an active site with a motif of known catalytic function and compares to proteins known in the database. Among the proteins with the highest alignment (lowest RMSD) with a RMSD of 1.17 is 2LPR, a hydrolase. | + | [[Image:2QRU(green)2LPR(yellow).png | thumb]]Analysis with Sprite displayed the proteins closest in structure to 2QRU. To do this, Sprite analyzed 2QRU for an active site with a motif of known catalytic function and compares to proteins known in the database. Among the proteins with the highest alignment (lowest RMSD) with a RMSD of 1.17 is 2LPR, a hydrolase. The amino acids included in the active site was 102 Ser, 104 Gly, 219 Asp, 247 His. |
'''Chimera''' | '''Chimera''' | ||
| Line 30: | Line 32: | ||
Investigation with InterPro showed the likely family of this enzyme was an alpha/beta hydrolase. Specifically, 2QRU was classified as a GDXG lipolytic enzyme. This family is generally associated with the hydrolysis of short fatty esters up to 8 carbons in length. Enzymes within this family are known to be involved in plant wall degradation and tryptophan synthesis. | Investigation with InterPro showed the likely family of this enzyme was an alpha/beta hydrolase. Specifically, 2QRU was classified as a GDXG lipolytic enzyme. This family is generally associated with the hydrolysis of short fatty esters up to 8 carbons in length. Enzymes within this family are known to be involved in plant wall degradation and tryptophan synthesis. | ||
| + | |||
'''SwissDock''' | '''SwissDock''' | ||
Current revision
Overview
| |||||||||||