Sandbox Aryan 20221057 BI3323-Aug2025
From Proteopedia
| (13 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | # | + | == Structure Overview == |
| + | |||
| + | PDB '''8YNY''' (4.52 Å '''cryo-EM''', EMDB: '''EMD-39431''') captures '''Cas9-sgRNA ribonucleoprotein''' in '''post-cleavage ternary complex''' with '''Widom 601 nucleosome'''.<ref name="pdb8yny">RCSB PDB - 8YNY</ref><ref name="nature">Nagamura R, et al. Structural insights into how Cas9 targets nucleosomes. Nat Commun. 2024</ref> | ||
| + | |||
| + | |||
| + | === Components === | ||
| + | - '''Cas9''' (Chain A): '''HNH/REC2 domains disordered''', '''bridge helix absent''' | ||
| + | - '''sgRNA''' (Chain B): Guides PAM1 targeting | ||
| + | - '''Nucleosome''': '''Histone octamer''' (H2A/H2B/H3/H4) + '''147 bp Widom601 DNA''' | ||
| + | - '''DNA state''': '''~15 bp unwrapped''' from histone octamer at '''PAM1 site''' | ||
| + | <Structure load='8YNY' size='350' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> | ||
| + | == Summary == | ||
| + | PDB '''8YNY''' (4.52 Å cryo-EM, EMDB: EMD-39431) captures '''Cas9-sgRNA ribonucleoprotein''' in '''post-cleavage complex''' with Widom 601 nucleosome.<ref name="pdb8yny"/> Cas9 targets '''linker DNA''' (PAM1/PAM28 sites) and '''entry-exit regions''' (SHL6), unwrapping ~15 bp DNA from histone octamer while avoiding tightly wrapped '''core DNA''' (SHL0-5).<ref name="nature">Nagamura R, et al. Structural insights into how Cas9 targets nucleosomes. Nat Commun. 2024</ref> | ||
| + | |||
| + | == Structure Overview == | ||
| + | '''Native-PAGE analysis''' confirms Cas9 preferentially cleaves '''nucleosome DNA ends''' where transient '''DNA breathing''' occurs, exploiting spontaneous unwrapping from histone octamer.<ref name="nature"/> Post-cleavage state reveals '''HNH/REC2 domains disordered''', '''bridge helix absent''', and both '''target/non-target DNA strands cleaved'''—consistent with binary biochemical assays.<ref name="nature"/> | ||
| + | |||
| + | [[8yny_scene1:1]] Interactive 3D overview (community scene + student analysis) | ||
| + | |||
| + | == PI Domain Interactions == | ||
| + | Cas9 '''PI domain''' (residues ~1100-1368) establishes '''multiple contacts''' with nucleosome: | ||
| + | - '''Histone tails''' (H2A/H3): Weak '''electrostatic interactions''' (acidic PI loop + basic tails) — '''non-essential''' for binding<ref name="nature"/> | ||
| + | - '''K1155''' (PI edge): '''Stabilizes post-cleavage complex''' via DNA phosphate backbone | ||
| + | - '''Core DNA loops''' ('''H1264/R1298/K1300'''): '''Inhibitory nonspecific binding''' to SHL0-5 DNA | ||
| + | |||
| + | **Mutagenesis validation**: '''H1264A/R1298Q/K1300A triple mutant''' ↑ nucleosome binding '''2-fold''' + cleavage efficiency '''3-5 fold''' (in vitro assays + rice callus genome editing).<ref name="nature"/> | ||
| + | |||
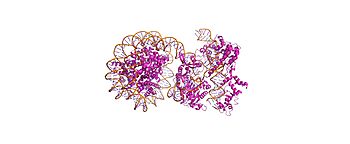
| + | [[Image:8YNYOverall.jpg|350px|thumb|1. Overall ternary complex - ~15 bp DNA unwrapping]] | ||
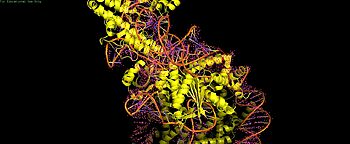
| + | [[Image:8YNYPI.jpg|350px|thumb|2. PI domain contacts - key residues highlighted]] | ||
| + | |||
| + | == Dual Inhibition Mechanism == | ||
| + | Nucleosomes inhibit Cas9 via '''two sequential barriers''': | ||
| + | |||
| + | # '''Access barrier''' (Stage 1): '''DNA end inflexibility''' at SHL0-5 prevents Cas9 binding to embedded PAM sites. '''Nucleosome breathing''' (transient unwrapping) + '''chromatin remodelers''' (SNF2h/RSC) enable access.<ref name="isaac">Isaac RS, et al. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. eLife. 2016</ref> | ||
| + | |||
| + | # '''Motion restriction''' (Stage 2): '''PI-core DNA trapping''' (H1264/R1298/K1300) physically '''stabilizes post-cleavage complex''', preventing '''HNH/RuvC domain rearrangements''' required for product release.<ref name="nature"/> | ||
| + | |||
| + | '''Entry/exit asymmetry''' from '''Widom601 sequence flexibility''' explains variable editing efficiency across chromatin contexts.<ref name="nature"/> | ||
| + | |||
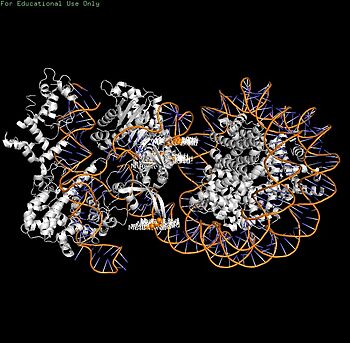
| + | [[Image:8YNYMutant.jpg|350px|thumb|3. Mutant sites (orange spheres): H1264/R1298/K1300]] | ||
| + | |||
| + | == Biological Context == | ||
| + | **Prokaryotic Cas9 in eukaryotic chromatin**: Reveals '''natural adaptation limitations''' + identifies '''chromatin-optimized variants''' (PI mutants) for therapeutic genome editing. '''Sequence-dependent nucleosome positioning''' + '''PTMs''' modulate endogenous Cas9 efficiency.<ref name="nature"/><ref name="isaac"/> | ||
| + | |||
| + | == Experimental Validation == | ||
| + | * '''Cryo-EM''': 4.52 Å resolution, captures '''DNA-attached state''' with fluctuating Cas9-nucleosome proximity<ref name="nature"/> | ||
| + | * '''Native-PAGE''': Quantifies '''position-dependent cleavage''' (PAM1 >> PAM14/17) | ||
| + | * '''Mutagenesis''': '''Triple PI mutant''' rescues inhibition '''in vitro/in vivo''' | ||
| + | |||
| + | [[8yny_scene2:1]] PI domain interactive 3D focus | ||
| + | |||
| + | == References == | ||
| + | <references /> | ||
| + | |||
| + | BI3323-Aug2025 | ||
| + | *Sandbox: proteopedia.org/wiki/Sandbox_BI3323_8YNY | ||
| + | *3D scenes: proteopedia.org/wiki/8yny (community resource) | ||
| + | *PNGs + 850-word analysis: Original PyMOL renders + primary literature synthesis | ||
| + | [[Category:CRISPR]] [[Category:Nucleosomes]] [[Category:Genome Editing]] | ||
Current revision
Contents |
Structure Overview
PDB 8YNY (4.52 Å cryo-EM, EMDB: EMD-39431) captures Cas9-sgRNA ribonucleoprotein in post-cleavage ternary complex with Widom 601 nucleosome.[1][2]
Components
- Cas9 (Chain A): HNH/REC2 domains disordered, bridge helix absent - sgRNA (Chain B): Guides PAM1 targeting - Nucleosome: Histone octamer (H2A/H2B/H3/H4) + 147 bp Widom601 DNA - DNA state: ~15 bp unwrapped from histone octamer at PAM1 site
|
Summary
PDB 8YNY (4.52 Å cryo-EM, EMDB: EMD-39431) captures Cas9-sgRNA ribonucleoprotein in post-cleavage complex with Widom 601 nucleosome.[1] Cas9 targets linker DNA (PAM1/PAM28 sites) and entry-exit regions (SHL6), unwrapping ~15 bp DNA from histone octamer while avoiding tightly wrapped core DNA (SHL0-5).[2]
Structure Overview
Native-PAGE analysis confirms Cas9 preferentially cleaves nucleosome DNA ends where transient DNA breathing occurs, exploiting spontaneous unwrapping from histone octamer.[2] Post-cleavage state reveals HNH/REC2 domains disordered, bridge helix absent, and both target/non-target DNA strands cleaved—consistent with binary biochemical assays.[2]
8yny_scene1:1 Interactive 3D overview (community scene + student analysis)
PI Domain Interactions
Cas9 PI domain (residues ~1100-1368) establishes multiple contacts with nucleosome: - Histone tails (H2A/H3): Weak electrostatic interactions (acidic PI loop + basic tails) — non-essential for binding[2] - K1155 (PI edge): Stabilizes post-cleavage complex via DNA phosphate backbone - Core DNA loops (H1264/R1298/K1300): Inhibitory nonspecific binding to SHL0-5 DNA
- Mutagenesis validation**: H1264A/R1298Q/K1300A triple mutant ↑ nucleosome binding 2-fold + cleavage efficiency 3-5 fold (in vitro assays + rice callus genome editing).[2]
Dual Inhibition Mechanism
Nucleosomes inhibit Cas9 via two sequential barriers:
- Access barrier (Stage 1): DNA end inflexibility at SHL0-5 prevents Cas9 binding to embedded PAM sites. Nucleosome breathing (transient unwrapping) + chromatin remodelers (SNF2h/RSC) enable access.[3]
- Motion restriction (Stage 2): PI-core DNA trapping (H1264/R1298/K1300) physically stabilizes post-cleavage complex, preventing HNH/RuvC domain rearrangements required for product release.[2]
Entry/exit asymmetry from Widom601 sequence flexibility explains variable editing efficiency across chromatin contexts.[2]
Biological Context
Experimental Validation
- Cryo-EM: 4.52 Å resolution, captures DNA-attached state with fluctuating Cas9-nucleosome proximity[2]
- Native-PAGE: Quantifies position-dependent cleavage (PAM1 >> PAM14/17)
- Mutagenesis: Triple PI mutant rescues inhibition in vitro/in vivo
8yny_scene2:1 PI domain interactive 3D focus
References
- ↑ 1.0 1.1 RCSB PDB - 8YNY
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 Nagamura R, et al. Structural insights into how Cas9 targets nucleosomes. Nat Commun. 2024
- ↑ 3.0 3.1 Isaac RS, et al. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. eLife. 2016
BI3323-Aug2025
- Sandbox: proteopedia.org/wiki/Sandbox_BI3323_8YNY
- 3D scenes: proteopedia.org/wiki/8yny (community resource)
- PNGs + 850-word analysis: Original PyMOL renders + primary literature synthesis