Arabidopsis thaliana PIN-FORMED 3 (AtPIN3)
From Proteopedia
| (9 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | ==AtPIN3== | ||
<span id="Figure_1:_Results"></span> | <span id="Figure_1:_Results"></span> | ||
[[Image:PIN3.png|thumb|right|250px|Figure 1: Polar Auxin Transport. Image by Jen Valenzuela (CC-BY-NC)<ref name="Ha" />.]] | [[Image:PIN3.png|thumb|right|250px|Figure 1: Polar Auxin Transport. Image by Jen Valenzuela (CC-BY-NC)<ref name="Ha" />.]] | ||
| Line 11: | Line 10: | ||
== Function == | == Function == | ||
Auxin and hence the PIN proteins are involved in many processes like '''embryogenesis, organogenesis, cell fate determination''', and '''cell division'''. It also contributes to '''trophic responses''' like gravitropism and phototropism<ref name="Su" />. | Auxin and hence the PIN proteins are involved in many processes like '''embryogenesis, organogenesis, cell fate determination''', and '''cell division'''. It also contributes to '''trophic responses''' like gravitropism and phototropism<ref name="Su" />. | ||
| + | |||
'''Mutation''' of PIN genes or their '''improper localization''' may lead to many '''developmental defects''' like shorter roots, reduced number of lateral roots, root meristem collapse, defective columella cells, abnormal cotyledons and altered leaf venation<ref name="Su" />. | '''Mutation''' of PIN genes or their '''improper localization''' may lead to many '''developmental defects''' like shorter roots, reduced number of lateral roots, root meristem collapse, defective columella cells, abnormal cotyledons and altered leaf venation<ref name="Su" />. | ||
| Line 16: | Line 16: | ||
<StructureSection load='7wks' size='340' side='right' caption='apo state' scene=''> | <StructureSection load='7wks' size='340' side='right' caption='apo state' scene=''> | ||
== Structure == | == Structure == | ||
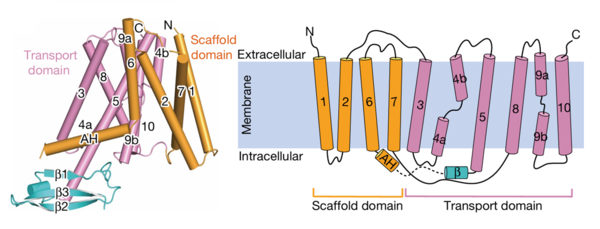
| - | AtPIN3 is its <scene name='10/1096831/Apo_state_of_pin3/1'>apo state</scene> is a '''homodimer''' with '''10 transmembrane (TM1-TM10) domains''' each [[#Figure_2:_Results|(Figure 2)]]. Both the N and the C terminal of the protein lie on the extracellular side. The 10 TM domains are divided into two groups a '''scaffold domain''' (TM1–2 and 6–7) and a '''transport domain''' (TM3–5 and 8–10)<ref name="Su" />. | + | AtPIN3 is its <scene name='10/1096831/Apo_state_of_pin3/1'>apo state</scene>(7wks) is a '''homodimer''' with '''10 transmembrane (TM1-TM10) domains''' each [[#Figure_2:_Results|(Figure 2)]]. Both the N and the C terminal of the protein lie on the extracellular side. The 10 TM domains are divided into two groups a '''scaffold domain''' (TM1–2 and 6–7) and a '''transport domain''' (TM3–5 and 8–10)<ref name="Su" />. |
<span id="Figure_2:_Results"></span> | <span id="Figure_2:_Results"></span> | ||
| Line 25: | Line 25: | ||
<span id="Figure_3:_Results"></span> | <span id="Figure_3:_Results"></span> | ||
| - | [[Image:PIN4.gif|400px|thumb|left|Figure 3:A clip taken from the YouTube video entitled: Structure and mechanism of the plant PIN-FORMED auxin transporter Posted by NanionTechnologies<ref> | + | [[Image:PIN4.gif|400px|thumb|left|Figure 3:A clip taken from the YouTube video entitled: "Structure and mechanism of the plant PIN-FORMED auxin transporter" Posted by NanionTechnologies.<ref>Posted by NanionTechnologies Speakers:Bjørn Panyella Pedersen and Ulrich Hammes. (2022). Structure and Mechanism of the plat PIN-FORMED auxin transporter. Retrieved from YouTube: https://www.youtube.com/watch?v=NPQK7T_UhrI</ref> Time Stamp: 30:00 to 30:15 |
| - | + | ]] | |
| - | + | ||
| - | + | The transport domain is predicted to undergo '''up-down rigid-body motion''' in an '''elevator-like model''' [[#Figure_3:_Results|(Figure 3)]]. Two weak helices TM4 and TM9 break in the middle and cross and connect to each other as short loops. These may provide a '''substrate binding site''' and allow for '''confirmational changes''' during auxin transport. A solvent accessible pathway is present between the scaffold and the transport domain. This was suggested as the location for the <scene name='10/1096831/Iaa_bound_state/2'>binding of IAA</scene>(7xxb). The elevator model is supported by a structural alignment of PIN3 in its apo and IAA bound state, which shows a movement of the transport domain 2-3 Å towards the scaffold domain once IAA binds<ref name="Su" />.[[#Figure_4:_Results|(Figure 4)]]. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | The transport domain is predicted to undergo '''up-down rigid-body motion''' in an '''elevator-like model''' [[#Figure_3:_Results|(Figure 3)]]. Two weak helices TM4 and TM9 break in the middle and cross and connect to each other as short loops. These may provide a '''substrate binding site''' and allow for '''confirmational changes''' during auxin transport. A solvent accessible pathway is present between the scaffold and the transport domain. This was suggested as the location for the <scene name='10/1096831/Iaa_bound_state/2'>binding of IAA</scene>. The elevator model is supported by a structural alignment of PIN3 in its apo and IAA bound state, which shows a movement of the transport domain 2-3 Å towards the scaffold domain once IAA binds<ref name="Su" />.[[#Figure_4:_Results|(Figure 4)]]. | ||
<span id="Figure_4:_Results"></span> | <span id="Figure_4:_Results"></span> | ||
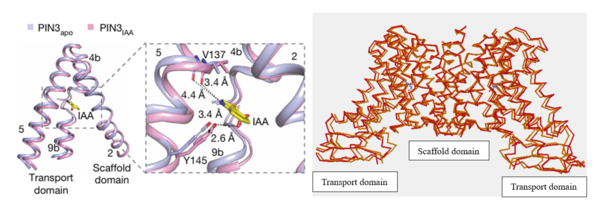
| - | [[Image:PIN2.png|600px|thumb|right|Figure 4: | + | [[Image:PIN2.png|600px|thumb|right|Figure 4:On the left, the local confirmation change observed between the structures of PIN3apo and PIN3IAA showing the change of state due to binding of IAA.Figure obtained from: Su, et. al.(2022)<ref name="Su" />Figure 2I. |
| - | + | On the right, an overall confirmation change observed in the transport domain created using TopMatch <ref>Sippl, & Wiederstein. (2020). Retrieved from TopMatch: https://topmatch.services.came.sbg.ac.at/index_jsmol.html?query=7xxb&qname=7xxb&target=7wks&tname=7wks</ref>]] | |
The protein has a '''cytosolic domain''' which is a cytosolic extension of <scene name='10/1096831/Apo_state_of_pin3/2'>TM5</scene>. It contains an '''Amphiphilic helix''' (AH) and '''3 beta-strands'''(β1-3). The loop between the AH and β3 has many phosphorylation sites that regulate the subcellular localization and transport activity of the protein<ref name="Su" />. | The protein has a '''cytosolic domain''' which is a cytosolic extension of <scene name='10/1096831/Apo_state_of_pin3/2'>TM5</scene>. It contains an '''Amphiphilic helix''' (AH) and '''3 beta-strands'''(β1-3). The loop between the AH and β3 has many phosphorylation sites that regulate the subcellular localization and transport activity of the protein<ref name="Su" />. | ||
| Line 45: | Line 39: | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | |||
| + | This web page was created for an assignment in Course BI3323-Aug2025 (Structural Biology), IISER, Pune | ||
Current revision
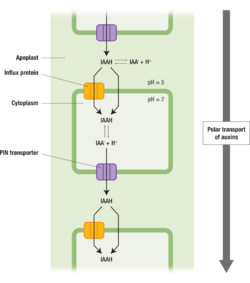
PIN-FORMED (PIN)[1] proteins in plants are responsible for the polar transport [2] of plant hormone auxin alongside AUXIN TRANSPORTER PROTEIN 1 (AUX1) and ATP-BINDING CASSETTE (ABC) transporters[2]. The polar transport of Auxin is crucial for proper plant growth and development. Auxin (IAAH) enters the cell through influx transporter or passes directly through the plasma membrane into the cytosol. Auxin dissociates to release a proton (H+) and anion (IAA-) in the cytoplasm due to higher pH. Due to its charge, it requires PIN proteins to carry it out of the cell. Once it reenters the apoplast it can bind to H again and it moves to the next cell[1] (Figure 1).
In Arabidopsis thaliana There are 8 PIN proteins divided into two subfamilies – six long PINs (PIN1-PIN4, PIN6 and PIN7) and two short PINs (PIN 5 and PIN8) that localize in the plasma membrane and the endoplasmic reticulum respectively. AtPIN3 is a long PIN that shares at least 54% similarity with other long PINs[2].
Function
Auxin and hence the PIN proteins are involved in many processes like embryogenesis, organogenesis, cell fate determination, and cell division. It also contributes to trophic responses like gravitropism and phototropism[2].
Mutation of PIN genes or their improper localization may lead to many developmental defects like shorter roots, reduced number of lateral roots, root meristem collapse, defective columella cells, abnormal cotyledons and altered leaf venation[2].
| |||||||||||
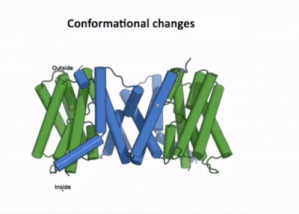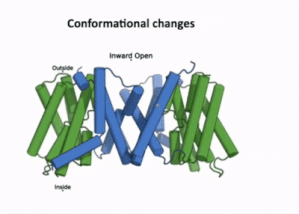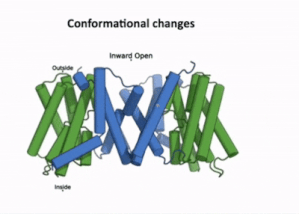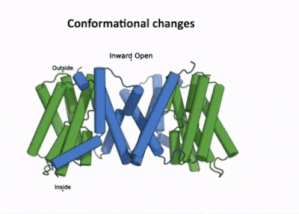
The transport domain is predicted to undergo up-down rigid-body motion in an elevator-like model (Figure 3). Two weak helices TM4 and TM9 break in the middle and cross and connect to each other as short loops. These may provide a substrate binding site and allow for confirmational changes during auxin transport. A solvent accessible pathway is present between the scaffold and the transport domain. This was suggested as the location for the (7xxb). The elevator model is supported by a structural alignment of PIN3 in its apo and IAA bound state, which shows a movement of the transport domain 2-3 Å towards the scaffold domain once IAA binds[2].(Figure 4).
The protein has a cytosolic domain which is a cytosolic extension of . It contains an Amphiphilic helix (AH) and 3 beta-strands(β1-3). The loop between the AH and β3 has many phosphorylation sites that regulate the subcellular localization and transport activity of the protein[2].
References
- ↑ 1.0 1.1 Ha, M., Morrow, M., & Algiers, K. (n.d.). Auxin. Retrieved from LibreTexts Biology: https://bio.libretexts.org/Bookshelves/Botany/Botany_(Ha_Morrow_and_Algiers)/04%3A_Plant_Physiology_and_Regulation/4.04%3A_Hormones/4.4.01%3A_Auxin#title
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 Su, N., Zhu, A., Tao, X. et al. Structures and mechanisms of the Arabidopsis auxin transporter PIN3. Nature 609, 616–621 (2022).DOI:https://doi.org/10.1038/s41586-022-05142-w
- ↑ Posted by NanionTechnologies Speakers:Bjørn Panyella Pedersen and Ulrich Hammes. (2022). Structure and Mechanism of the plat PIN-FORMED auxin transporter. Retrieved from YouTube: https://www.youtube.com/watch?v=NPQK7T_UhrI
- ↑ Sippl, & Wiederstein. (2020). Retrieved from TopMatch: https://topmatch.services.came.sbg.ac.at/index_jsmol.html?query=7xxb&qname=7xxb&target=7wks&tname=7wks
This web page was created for an assignment in Course BI3323-Aug2025 (Structural Biology), IISER, Pune