We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Nitrogenase
From Proteopedia
(Difference between revisions)
| (156 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
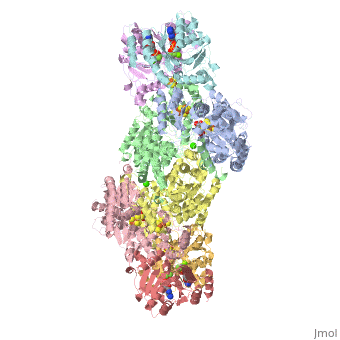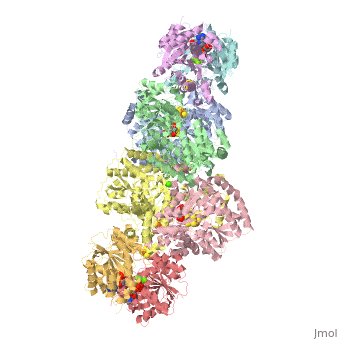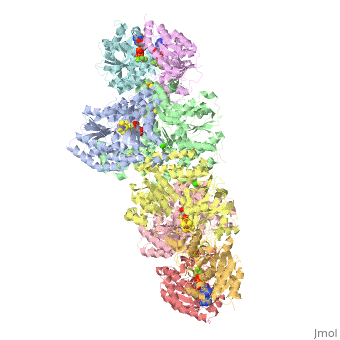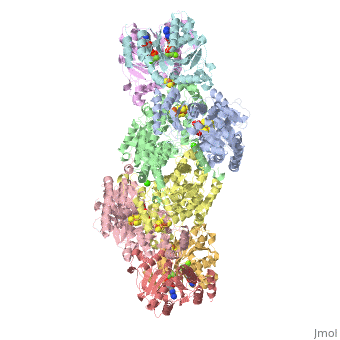
| - | + | <StructureSection load="1N2C" size="350" color="white" caption="Nitrogenase complex: α (grey and green) and β (pink and yellow) chains, nitrogenase iron protein ( purple,cyan, red and gold), showing Fe-Mo-S cluster complex with ADP, adipic acid, AlF4, Ca+2 and Mg+2 ions [[1n2c]]"> | |
| + | __TOC__ | ||
| + | ==Function== | ||
| - | + | '''Nitrogenase''' (Nase) is an enzyme that fixes atmospheric nitrogen (N<sub>2</sub>) into ammonia. Though abundantly present in the atmosphere, most organisms cannot utilize N<sub>2</sub> directly, and must instead take it in through other forms, like ammonia or nitrate. The triple bond in N<sub>2</sub> is highly resistant to changes in oxidation state, and nitrogenases, found only in nitrogen-fixing bacteria, are the only proteins capable of reducing N<sub>2</sub> to ammonia. | |
| - | This picture was created with RasMol. You can create similar pictures by clicking on the accession code above and then picking one of the options under View Structure. The chromophore is called "CRO" in this file, and it is residue number 66 in the protein chain. | ||
| + | Nitrogenase catalyzes the following reaction: | ||
| + | N<sub>2</sub> + 8 H<sup>+</sup> + 16 MgATP + 8 e<sup>-</sup> → 2NH<sub>3</sub> + H<sub>2</sub> + 16 MgADP + 16 P<sub>i</sub> | ||
| - | + | Two different proteins comprise the nitrogenase complex. The FeMo protein binds substrate and reduces H<sup>+</sup> and N<sub>2</sub> to H<sub>2</sub> and ammonia, while the Fe protein receives electrons from ferredoxin, hydrolyzes ATP, and reduces the FeMo protein. To the right is shown a crystal structure (PDB entry [[1n2c]]<ref>PMID:9163420</ref>) where two complexes of FeMo protein bound to Fe protein were crystallized together. Click here to see only <scene name='Sandbox_10/1n2c_single_complex/2'>one complex</scene>. | |
| + | |||
| + | |||
| + | The <scene name='Sandbox_10/1n2c_fe_protein/1'>Fe protein</scene> is here bound to two <scene name='Sandbox_10/1n2c_atp/3'>ADP x AlF4-</scene>, an analog for the planar transition state of ATP hydrolysis. The motif that binds ATP is a conserved nucleotide binding motif called Walker's motif A. Coloring by <scene name='Sandbox_10/1n2c_atp_evolutionary/1'>evolutionary conservation</scene>, the nucleotide binding pocket is clear. At the bottom of the protein, where the Fe protein comes into contact with the FeMo protein, is a <scene name='Sandbox_10/1n2c_fes_cluster_cys/1'>4Fe:4S cluster</scene>, held in place by cysteines. This cluster accepts electrons from ferredoxin and gives electrons to the FeMo protein. | ||
| + | |||
| + | |||
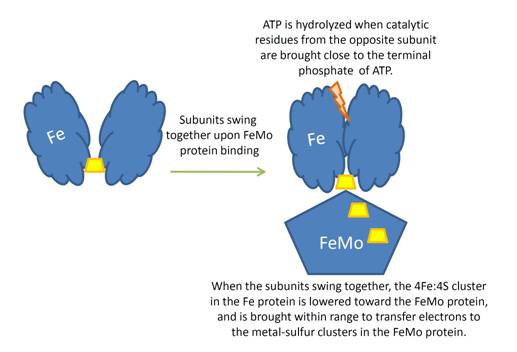
| + | When the Fe protein is bound to the FeMo protein (<scene name='Sandbox_10/1n2c_single_complex/2'>zoom out</scene>), ATP is hydrolyzed and electrons that were transferred to the 4Fe:4S cluster of the Fe protein by ferredoxin are transferred to the FeMo protein. The crystal structure of the complete nitrogenase complex reveals how the binding of the Fe and FeMo proteins, the hydrolysis of ATP, and the transfer of electrons are all coupled. The figure below summarizes how these processes are linked. | ||
| + | |||
| + | [[Image:clip_image002.jpg]] | ||
| + | |||
| + | Before this crystal structure showing the complete nitrogenase complex was solved, crystal structures of the Fe and FeMo proteins had been solved, but the mechanisms of ATP hydrolysis and electron transfer were still unknown. In this structure, as indicated in the figure above, the two subunits of the Fe protein were observed to have swung closer together. This movement results from Fe protein binding to FeMo protein. Focusing in on the <scene name='Sandbox_10/1n2c_atp/3'>ATP binding pocket</scene> discussed above, especially on the <scene name='Sandbox_10/1n2c_atp_lys_10/1'>AlF4-</scene> that is an analog for the negatively charged planar transition state reveals that there are several positive charges in this vicinity that stabilize the transition state. Importantly, Lys 10 from the opposite subunit is an important source of stabilizing positive charge. Only in this structure where the two subunits have swung closer together is this residue in position to help catalyze the hydrolysis of the terminal phosphate of ATP. | ||
| + | |||
| + | |||
| + | For these reasons, binding of Fe protein to FeMo protein results in hydrolysis of ATP. Additionally, the 4Fe:4S cluster is lowered close enough to the metal-sulfur clusters of the FeMo protein that electron transfer can occur. All three clusters found in the Fe protein-FeMo protein complex can be seen <scene name='Sandbox_10/1n2c_clusters/2'>here</scene>. Once the electrons have passed from the 4Fe:4S cluster of the Fe protein to the 8Fe:7S cluster of the FeMo protein, they then transfer to the 7Fe:Mo:9S:homocitrate:X cluster where X is an unidentified light atom. It is at this cluster where reduction of N<sub>2</sub> and H<sup>+</sup> occur. The exact mechanism of reduction, however, is still unknown. | ||
| + | |||
| + | ==3D structure of Nitrogenase== | ||
| + | [[Nitrogenase 3D structures]] | ||
| + | |||
| + | </StructureSection> | ||
| + | |||
| + | == References == | ||
| + | <references/> | ||
| + | |||
| + | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ Schindelin H, Kisker C, Schlessman JL, Howard JB, Rees DC. Structure of ADP x AIF4(-)-stabilized nitrogenase complex and its implications for signal transduction. Nature. 1997 May 22;387(6631):370-6. PMID:9163420 doi:10.1038/387370a0
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Eran Hodis, David Canner, Joel L. Sussman