Zinc Fingers
From Proteopedia
(→Exploring the Structure) |
|||
| (20 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| + | <StructureSection load='2CI3' size='400' side='right' caption='Zn stabilize the structure of Zinc Finger Domain (PDB code [[1zaa]])' scene='Zinc_Fingers/Zinc_finger_initial/3'> | ||
| + | |||
[[Image:1ZAA.jpg |left |thumb |Cartoon representation for Zinc Finger [[1zaa]] with Zn atoms (green)]] | [[Image:1ZAA.jpg |left |thumb |Cartoon representation for Zinc Finger [[1zaa]] with Zn atoms (green)]] | ||
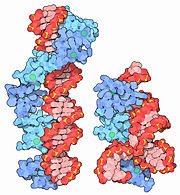
| + | [[Image:MotM 1tf6-1un6.jpg |left |thumb | left: [[1tf6]] DNA (red) with 6 zinc fingers (blue). right:[[1un6]] and [[2hgh]] ribosomal RNA (red) with 3 zinc fingers (blue). Figure Credit: [[Molecule of the Month]]]] | ||
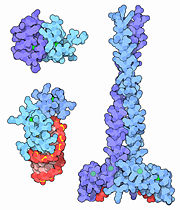
| - | + | [[Image:MotM Zincfingers.jpg |right |thumb |upper left [[1y0j]], lower left [[1a1t]], right [[1joc]]. Figure Credit: [[Molecule of the Month]]]] | |
| - | + | ||
| - | + | == Overview == | |
| - | <scene name=' | + | Few classes of transcription factors are as significant as the '''zinc fingers'''. While there are zinc-containing subregions in other proteins, the distinguishing feature of a zinc finger is the spontaneous folding process which is facilitated by a <scene name='10/100440/Zincs_labeled/1'>zinc ion</scene>. Since the zinc fingers are so small, they cannot rely on the collective strength of many hydrogen bonds or hydrophobic interactions to drive their folding. They are highly specific to their sequence and yet are able to target sequences common to multiple loci for regulation of a common function that may be influenced by many genes. They can also serve as a carrier for another domain which may bind covalently to the DNA, effecting permanent shutdown of a gene’s expression. Attesting to their importance in the human genome, a family of over 700 human proteins contain a zinc finger domain, a number exceeded only the immunoglobins, and then only slightly. |
| - | == | + | == Basic Structural Profile of a Zinc Finger == |
| + | The simplest group of zinc fingers, referred as [http://proteopedia.org/wiki/index.php/1znf C<sub>2</sub>H<sub>2</sub>] zinc fingers, consists of <scene name='10/100440/2ndary_structure/1'>two antiparallel β-pleated sheets and a right-handed α-helix</scene>. The name [http://proteopedia.org/wiki/index.php/1znf C<sub>2</sub>H<sub>2</sub>], or Cis<sub>2</sub>His<sub>2</sub>, gives a nod to the <scene name='10/100440/His_cys_zn/1'>two cysteine and two histidine</scene> residues involved in coordination of the zinc ion. The turn between the two β-pleated sheets forms a hydrophobic pocket near where the zinc ion is bound. Typically, the hydrophobic pocket is formed as a result of interactions between <scene name='10/100440/His_cys_zn_phe/1'>phenylalanines</scene> and leucines in close proximity. While both the coordination of the zinc ligand and the presence of the hydrophobic pocket stabilize the small zinc finger domain, the zinc ligand is responsible for a majority of the stability imparted to the motif. Because cells contain a highly reducing environment, sulfide bridges are unable to stabilize small protein domains. With a single oxidation state and the ability to accommodate both nitrogen and sulfur, Zinc is an ideal stabilizer. Due to the structural stability imparted to the motif by the zinc ion, zinc fingers are considerably smaller than most other proteins, usually ranging between 25 and 30 amino acids in length. | ||
| + | As a corollary of their small size, they are extremely agile and mobile, and so are the genes that encode them. Thus, they are easily able to bind to DNA, among other substrates, in part due to interactions of <scene name='10/100440/Po4-arg/1'>positively charged residues</scene> with the negatively charged phosphate backbone. By slipping into the major groove of DNA, they are able to use their <scene name='10/100440/Dna-protein_interaction/1'>amino acids</scene> to check for proper base identity. This allows the zinc fingers to bind to sequences normally unavailable to other, larger DNA-binding structural motifs. | ||
| - | + | As a family, the structure of the zinc fingers is as polymorphous as it is unique. Some are coordinated primarily or exclusively by cysteine, and many form much more complex structures than the β-hairpin of Cis<sub>2</sub>His<sub>2</sub> types. | |
| - | == | + | ==Example of a zinc finger: Gal4== |
| - | + | <scene name='10/100440/Gal4/1'>Gal4</scene> is a positive regulator of galactose-induced genes, and is used in yeast two hybrid systems. The N terminal fragment of Gal4 binds as a dimer to a symmetrical 17-base-pair sequence. Each subunit folds into 3 distinct modules: A compact, <scene name='10/100440/Metal_binding_domain/1'>metal binding domain</scene>, and an extended <scene name='10/100440/Linker/1'>linker</scene> (41-49), and an alpha-helical <scene name='10/100440/Gal4_dimerization/1'>dimerization</scene> element (50-64). A small, Zn(2+)-containing domain recognizes a conserved <scene name='10/100440/Gal4_ccg/1'>CCG</scene> triplet at each end of the site through <scene name='10/100440/Gal4_ccg_lys/1'>direct contacts</scene> with the major groove. The relatively open structure of the complex would allow another protein to bind coordinately with GAL4. | |
| - | + | ||
| - | == | + | == Biological Role and Regulation == |
| + | Given the prevalence of the zinc fingers in nature, it is no surprise to learn that they are key players in many biological processes. Gli-1, a zinc-finger transcription factor involved in embryonic development, is also implicated in carcinogenesis when it is over-expressed in dividing cells. Likewise, Gli-3 is another zinc-finger transcription factor whose shortened repressor form has roles in modulating regulation of genes controlling apoptosis, and consequently has a dramatic effect on development of the limb bud and the digit formation that follows. In part, the regulation of the zinc fingers is kept in check by other repressors or activators, as well as various types of chromatin remodeling (e.g. acetylation, methylation) which definitively blocks transcription of their targets. | ||
| - | + | == Clinical Applications == | |
| + | The advantages of using ZFPs in a clinical setting are numerous, the least of which is not their ability to bind DNA. As a result, the ZFP must bind two only two copies of its target, as opposed to therapeutics directed at mRNA, for example, which have to are in direct quantitative competition with their target. Because of this unique advantage as well as their modularity and ability to work in clusters, the ZFPs are currently the subject of extensive research for use in genetic therapy. Their versatility and modularity among targets can be explained as a function of their α-helical sidechains, which they use to interact electrostatically with multiple sequences. Because they can withstand multiple mutations without losing their functional structure, they make great candidates for specific gene-targeting applications. In theory, the zinc finger as a therapeutic agent could unilaterally control gene expression, given that a transcription factor had been synthesized which possessed the proper sequence identity for binding to the gene or the regulated protein. | ||
| - | == | + | </StructureSection> |
| - | + | == 3D Structure of Zinc Finger Domains == | |
| - | Zinc | + | [[1hvo]] - HIV-1 nucleocpasid<br /> |
| + | [[1x68]] - FHL5<br /> | ||
| + | [[1x6a]] - LIM kinase 2<br /> | ||
| + | [[1x6f]] - Zinc finger protein 462<br /> | ||
| + | [[2bai]] - Genome polyprotein<br /> | ||
| + | [[2ctu]] - Zinc finger protein 483<br /> | ||
| + | [[2ds6]] - [[Clp Protease]]<br /> | ||
| + | [[2e72]] - Pogo transposable element<br /> | ||
| + | [[2epc]] - Zinc finger protein 32<br /> | ||
| + | [[2epp]] - Zinc finger protein 278<br /> | ||
| + | [[2epq]] - Zinc finger protein 278<br /> | ||
| + | [[2epr]] - Zinc finger protein 278<br /> | ||
| + | [[2eps]] - Zinc finger protein 278<br /> | ||
| + | [[2ept]] - Zinc finger protein 32<br /> | ||
| + | [[2epu]] - Zinc finger protein 32<br /> | ||
| + | [[2epv]] - Zinc finger protein 268<br /> | ||
| + | [[2epw]] - Zinc finger protein 268<br /> | ||
| + | [[2epx]] - Zinc finger protein 28<br /> | ||
| + | [[2epy]] - Zinc finger protein 268<br /> | ||
| + | [[2epz]] - Zinc finger protein 28<br /> | ||
| + | [[2eq0]] - Zinc finger protein 347<br /> | ||
| + | [[2eq1]] - Zinc finger protein 347<br /> | ||
| + | [[2eq2]] - Zinc finger protein 347<br /> | ||
| + | [[2eq3]] - Zinc finger protein 347<br /> | ||
| + | [[2eq4]] - Zinc finger protein 224<br /> | ||
| + | [[2rpp]] - Muscleblind-like protein 2<br /> | ||
| + | [[2ysv]] - Zinc finger protein 473<br /> | ||
| - | == Exploring the Structure == | ||
| - | <applet load='1znf' scene='Zinc_Fingers/Zinc_fingers/2' size='400' frame='true' align='right' /> | ||
| - | When you go to the PDB to explore zinc fingers, try not to be overwhelmed: there are currently over a thousand structures containing all sorts of zinc fingers, knuckles, treble clefs, ribbons, and other fanciful folds. [[1znf]]. shows a classic zinc finger. Since it's such a small structure, with only | ||
| - | <scene name='Zinc_Fingers/Zinc_fingers/1'>25 amino acids</scene>, it's worth taking a closer look at what each part is doing. The <scene name='Zinc_Fingers/Zinc_fingers_cys/3'>two cysteines</scene> and <scene name='Zinc_Fingers/Zinc_fingers_his/4'>two histidines</scene>, arranged in a tetrahedron around the zinc. Just above them, <scene name='Zinc_Fingers/Zinc_fingers_phe/2'>a phenylalanine</scene> and <scene name='Zinc_Fingers/Zinc_fingers_leu/1'>a leucine</scene> form a tiny hydrophobic core inside the small folded chain. The rest of the amino acids all face more-or-less outwards, and can be used for various functional tasks. For DNA binding proteins, the amino acids at the top of the alpha helix are typically used for recognition. This is shown on the structure at right, [[1zaa]]. It contains <scene name='Zinc_Fingers/Zinc_fingers_1zaa_dna/3'>three linked zinc fingers</scene> bound to DNA. <scene name='Zinc_Fingers/Zinc_fingers_1zaa_dna/4'>The amino acids shown in red</scene> extend towards the DNA and read the base sequence. | ||
{{Clear}} | {{Clear}} | ||
| Line 37: | Line 64: | ||
# [http://en.wikipedia.org/wiki/Zinc_finger Zinc Finger in Wikipedia] | # [http://en.wikipedia.org/wiki/Zinc_finger Zinc Finger in Wikipedia] | ||
# [http://www.zincfingers.org/ Zinc Finger Consortium] | # [http://www.zincfingers.org/ Zinc Finger Consortium] | ||
| + | # [http://mgl.scripps.edu/people/goodsell/pdb/pdb87/pdb87_1.html Molecule of the Month on Zinc Fingers] by David Goodsell | ||
| + | #''Molecule of the Month'' at [[Teaching Scenes, Tutorials, and Educators' Pages]] | ||
| + | # For Additional Information, See: [[Transcription]] <br /> | ||
== Content Donators == | == Content Donators == | ||
| - | + | ||
| + | Many images on this page are the work of David S. Goodsell, who has given permission for their inclusion in [[Proteopedia]]: | ||
* Content adapted with permission from David S. Goodsell's [http://mgl.scripps.edu/people/goodsell/pdb/pdb87/pdb87_1.html Molecule of the Month on Zinc Fingers] | * Content adapted with permission from David S. Goodsell's [http://mgl.scripps.edu/people/goodsell/pdb/pdb87/pdb87_1.html Molecule of the Month on Zinc Fingers] | ||
Current revision
| |||||||||||
3D Structure of Zinc Finger Domains
1hvo - HIV-1 nucleocpasid
1x68 - FHL5
1x6a - LIM kinase 2
1x6f - Zinc finger protein 462
2bai - Genome polyprotein
2ctu - Zinc finger protein 483
2ds6 - Clp Protease
2e72 - Pogo transposable element
2epc - Zinc finger protein 32
2epp - Zinc finger protein 278
2epq - Zinc finger protein 278
2epr - Zinc finger protein 278
2eps - Zinc finger protein 278
2ept - Zinc finger protein 32
2epu - Zinc finger protein 32
2epv - Zinc finger protein 268
2epw - Zinc finger protein 268
2epx - Zinc finger protein 28
2epy - Zinc finger protein 268
2epz - Zinc finger protein 28
2eq0 - Zinc finger protein 347
2eq1 - Zinc finger protein 347
2eq2 - Zinc finger protein 347
2eq3 - Zinc finger protein 347
2eq4 - Zinc finger protein 224
2rpp - Muscleblind-like protein 2
2ysv - Zinc finger protein 473
Additional Information
- Zinc Finger in Wikipedia
- Zinc Finger Consortium
- Molecule of the Month on Zinc Fingers by David Goodsell
- Molecule of the Month at Teaching Scenes, Tutorials, and Educators' Pages
- For Additional Information, See: Transcription
Content Donators
Many images on this page are the work of David S. Goodsell, who has given permission for their inclusion in Proteopedia:
- Content adapted with permission from David S. Goodsell's Molecule of the Month on Zinc Fingers
Proteopedia Page Contributors and Editors (what is this?)
Ala Jelani, Ann Taylor, Eran Hodis, Michal Harel, Tyler Combs, Joel L. Sussman, David Canner, Eric Martz