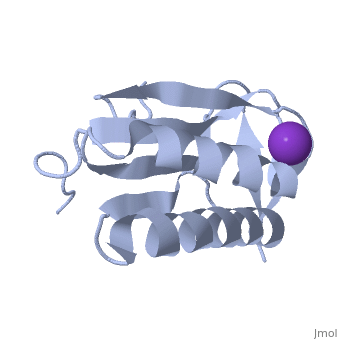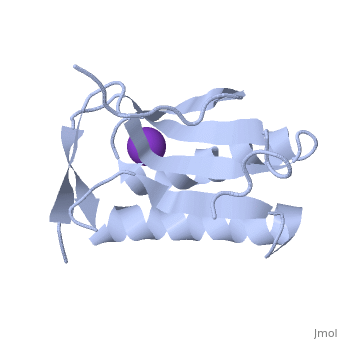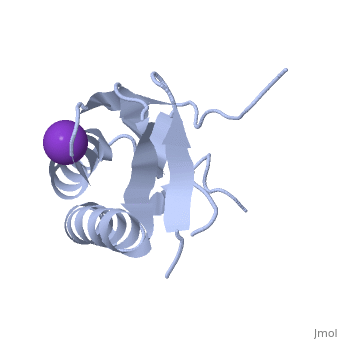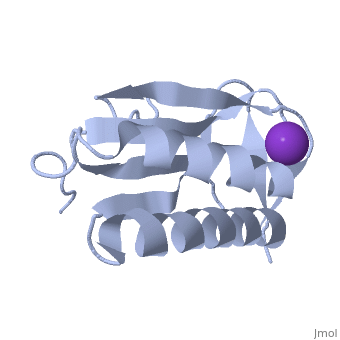
We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
2pa2
From Proteopedia
(Difference between revisions)
| (12 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | {{Seed}} | ||
| - | [[Image:2pa2.png|left|200px]] | ||
| - | < | + | ==Crystal structure of human Ribosomal protein L10 core domain== |
| - | + | <StructureSection load='2pa2' size='340' side='right'caption='[[2pa2]], [[Resolution|resolution]] 2.50Å' scene=''> | |
| - | You may | + | == Structural highlights == |
| - | + | <table><tr><td colspan='2'>[[2pa2]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2PA2 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2PA2 FirstGlance]. <br> | |
| - | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.5Å</td></tr> | |
| - | -- | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=K:POTASSIUM+ION'>K</scene></td></tr> |
| - | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2pa2 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2pa2 OCA], [https://pdbe.org/2pa2 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2pa2 RCSB], [https://www.ebi.ac.uk/pdbsum/2pa2 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2pa2 ProSAT], [https://www.topsan.org/Proteins/RSGI/2pa2 TOPSAN]</span></td></tr> | |
| + | </table> | ||
| + | == Disease == | ||
| + | [https://www.uniprot.org/uniprot/RL10_HUMAN RL10_HUMAN] Defects in RPL10 are a cause of susceptibility to autism X-linked type 5 (AUTSX5) [MIM:[https://omim.org/entry/300847 300847]. A complex multifactorial, pervasive developmental disorder characterized by impairments in reciprocal social interaction and communication, restricted and stereotyped patterns of interests and activities, and the presence of developmental abnormalities by 3 years of age. Most individuals with autism also manifest moderate mental retardation. Note=RPL10 is involved in autism only in rare cases. Two hypomorphic variants affecting the translation process have been found in families with autism spectrum disorders, suggesting that aberrant translation may play a role in disease mechanisms.<ref>PMID:16940977</ref> <ref>PMID:21567917</ref> | ||
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/RL10_HUMAN RL10_HUMAN] | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/pa/2pa2_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2pa2 ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | A phylogenetically conserved ribosomal protein L16p/L10e organizes the architecture of the aminoacyl tRNA binding site on the large ribosomal subunit. Eukaryotic L10 also exhibits a variety of cellular activities, and, in particular, human L10 is known as a putative tumor suppressor, QM. We have determined the 2.5-A crystal structure of the human L10 core domain that corresponds to residues 34-182 of the full-length 214 amino acids. Its two-layered alpha+beta architecture is significantly similar to those of the archaeal and bacterial homologues, substantiating a high degree of structural conservation across the three phylogenetic domains. A cation-binding pocket formed between alpha2 and beta 6 is similar to that of the archaeal L10 protein but appears to be better ordered. Previously reported L10 mutations that cause defects in the yeast ribosome are clustered around this pocket, indicating that its integrity is crucial for its role in L10 function. Characteristic interactions among Arg90-Trp171-Arg139 guide the C-terminal part outside of the central fold, implying that the eukaryote-specific C-terminal extension localizes on the outer side of the ribosome. | ||
| - | + | Crystal structure of human ribosomal protein L10 core domain reveals eukaryote-specific motifs in addition to the conserved fold.,Nishimura M, Kaminishi T, Takemoto C, Kawazoe M, Yoshida T, Tanaka A, Sugano S, Shirouzu M, Ohkubo T, Yokoyama S, Kobayashi Y J Mol Biol. 2008 Mar 21;377(2):421-30. Epub 2008 Jan 11. PMID:18258260<ref>PMID:18258260</ref> | |
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
| + | <div class="pdbe-citations 2pa2" style="background-color:#fffaf0;"></div> | ||
| - | + | ==See Also== | |
| - | + | *[[Ribosomal protein L10|Ribosomal protein L10]] | |
| - | + | == References == | |
| - | + | <references/> | |
| - | + | __TOC__ | |
| - | + | </StructureSection> | |
| - | == | + | |
| - | + | ||
| - | + | ||
| - | == | + | |
| - | + | ||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
| - | [[Category: Kaminishi | + | [[Category: Large Structures]] |
| - | [[Category: Kawazoe | + | [[Category: Kaminishi T]] |
| - | [[Category: Kobayashi | + | [[Category: Kawazoe M]] |
| - | [[Category: Nishimura | + | [[Category: Kobayashi Y]] |
| - | [[Category: Ohkubo | + | [[Category: Nishimura M]] |
| - | + | [[Category: Ohkubo T]] | |
| - | [[Category: Shirouzu | + | [[Category: Shirouzu M]] |
| - | [[Category: Sugano | + | [[Category: Sugano S]] |
| - | [[Category: Takemoto | + | [[Category: Takemoto C]] |
| - | [[Category: Tanaka | + | [[Category: Tanaka A]] |
| - | [[Category: Yokoyama | + | [[Category: Yokoyama S]] |
| - | [[Category: Yoshida | + | [[Category: Yoshida T]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Crystal structure of human Ribosomal protein L10 core domain
| |||||||||||
Categories: Homo sapiens | Large Structures | Kaminishi T | Kawazoe M | Kobayashi Y | Nishimura M | Ohkubo T | Shirouzu M | Sugano S | Takemoto C | Tanaka A | Yokoyama S | Yoshida T