This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Transcription Termination Factor Rho
From Proteopedia
(added missing residues and element color key) |
|||
| (6 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | {{STRUCTURE_1pvo| PDB=1pvo | SIZE=400| SCENE= |right|CAPTION=E. coli transcription termination factor Rho complex wit ssRNA and ANPPNP, [[1pvo]] }} | |
| + | |||
'''Transcription termination factor Rho''' is a ring-shaped RNA-DNA '''helicase''' that induces release of transcription complexes at specific loci of bacterial genomes. Several structures of Rho in various liganded states haven been solved by X-ray crystallography by the J. M. Berger lab (Berkeley): [[1a8v]], [[1pv4]], [[1pvo]], [[1xpr]], [[1xpu]], [[2a8v]], and [[2ht1]]. | '''Transcription termination factor Rho''' is a ring-shaped RNA-DNA '''helicase''' that induces release of transcription complexes at specific loci of bacterial genomes. Several structures of Rho in various liganded states haven been solved by X-ray crystallography by the J. M. Berger lab (Berkeley): [[1a8v]], [[1pv4]], [[1pvo]], [[1xpr]], [[1xpu]], [[2a8v]], and [[2ht1]]. | ||
| Line 11: | Line 12: | ||
{{Template:ColorKey_Element_O}} | {{Template:ColorKey_Element_O}} | ||
{{Template:ColorKey_Element_N}} | {{Template:ColorKey_Element_N}} | ||
| - | {{Template:ColorKey_Element_P}}). Also colored are | + | {{Template:ColorKey_Element_P}}). Also colored are |
| + | <font color='#00a600'>'''Lys181'''</font>, | ||
| + | <font color='#cc3700'>'''Lys283'''</font>, | ||
| + | <font color='#20603c'>'''Arg347'''</font>, | ||
| + | <font color='#974e94'>'''Arg353'''</font>, and (in black) '''background RNA'''. See also [[Transcription and RNA Processing]]. | ||
| + | |||
| + | ==3D structures of helicase== | ||
| + | |||
| + | [[Helicase]] | ||
| + | |||
| + | ==Additional Resources== | ||
| + | For additional information, see: [[Transcription and RNA Processing]] | ||
| + | <br /> | ||
==Notes== | ==Notes== | ||
<references /> | <references /> | ||
Current revision
Transcription termination factor Rho is a ring-shaped RNA-DNA helicase that induces release of transcription complexes at specific loci of bacterial genomes. Several structures of Rho in various liganded states haven been solved by X-ray crystallography by the J. M. Berger lab (Berkeley): 1a8v, 1pv4, 1pvo, 1xpr, 1xpu, 2a8v, and 2ht1.
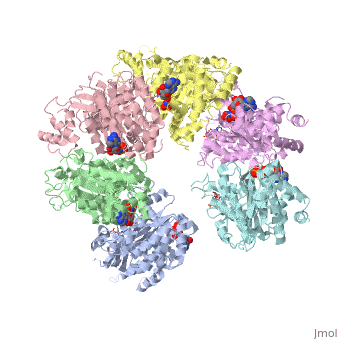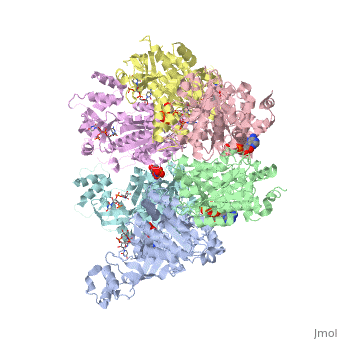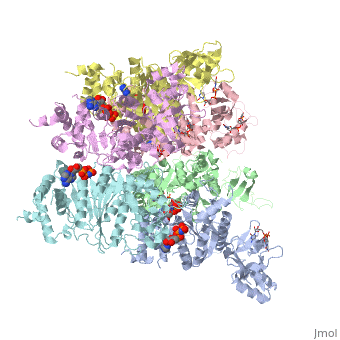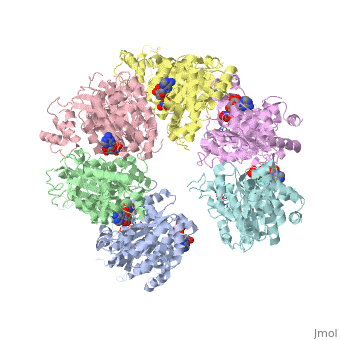
The asymmetric unit solved as 2ht1 contains only two chains, so it represents only one third of the biological unit, which is a homo-hexamer. The scene here shows this homo-hexamer in a closed state ()[1]. Note that this model has only the middle 324 amino acids in each chain, lacking the first 52 and the last 57 amino acids in each chain[2].
In this model (), the Protein hexamer is bound to RNA, and some incidental "Solvent" (sulfate ions).
Here is one representation showing the same Rho hexamer in a closed state and with . In this scene, the foreground RNA is colored by element (C O N P). Also colored are Lys181, Lys283, Arg347, Arg353, and (in black) background RNA. See also Transcription and RNA Processing.
3D structures of helicase
Additional Resources
For additional information, see: Transcription and RNA Processing
Notes
- ↑ File Image:Rho1.pdb derived from 2ht1 by symmetry operations
- ↑ Missing residues can be found by aligning the PDB file's SEQRES records with the sequence of the residues assigned atomic coordinates, as listed by the S2C Server