User:Grace Natalie
From Proteopedia
| (119 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | <font size= | + | <center><font size=6 face="Comic Sans MS">ATP Binding in Glutamine Synthetase</font></center> |
| + | <br><br> | ||
| + | <font size=4 face ="Arial">Background</font> | ||
| + | <br> | ||
| + | <applet load='1lgr' size='350' color='white' frame='true' align='right' caption='Glutamine Synthetase from Salmonella typhimurium'/> | ||
| + | <font size=2>Glutamine synthetase (GS) catalyzes the ATP-dependent condensation of ammonia and | ||
| + | glutamate to yield glutamine, ADP, and inorganic phosphate in the presence of divalent cations | ||
| + | <ref name="Liaw">Liaw, S-H, et.al.,Discovery of the ammonium substrate site on glutamine synthetase, a third cation binding site Protein Sci. 1995 4: 2358-2365</ref> . | ||
| + | The reaction occurs in two steps with γ-glutamyl phosphate as an intermediate and is used by | ||
| + | bacteria to introduce reduced nitrogen into cellular metabolism. GS is a dodecamer formed from | ||
| + | two face-to-face hexameric rings of subunits, with 12 active sites formed between monomers | ||
| + | <ref name="Book">Gill, H & Eisenberg, D., Biochemistry 2001 40: 1903-1912</ref> .</font> | ||
| + | <br><br> | ||
| + | <center><font size=4 face ="Arial">Overall Reaction of Glutamine Synthetase</font></center> | ||
| + | <center><font size=2>Glutamate + NH<SUB>4</SUB><SUP>+</SUP> + ATP --> glutamine + ADP + P<SUB>i</SUB></font></center> | ||
| + | <br> | ||
| + | <font size=4 face ="Arial">Overall Mechanism</font> | ||
| + | <br> | ||
| + | <font size=2>The first step is the formation of the activated intermediate γ-glutamyl phosphate. | ||
| + | The n2 ion coordinates the phosphate oxygens of ATP to allow phosphoryl transfer to the | ||
| + | γ-carboxylate group of glutatmate, yielding the intermediate <ref name="Eisenberg">D. Eisneberg et al / Biochimica et Biophysica Acta 1477 (2000) 124</ref> . The second step is the attack on the intermediate by ammonia | ||
| + | therefore releasing free phosphate to yield glutamine.</font> | ||
| + | <br> | ||
| + | <font size=4 face ="Arial">ATP binding site</font> | ||
| + | <br> | ||
| + | <font size=2>Each active site of GS is described as a 'bifunnel in which ATP and glutamate bind at opposite ends. | ||
| + | The ATP binding site is referred to as the top of the bifunnel because it opens to the external 6-fold surface of GS (figure below) <ref name="Eisenberg"/>. At the the joint of the <scene name='User:Grace_Natalie/Atp_binding_site/2'> bifunnel </scene> are two cation binding sites, n1 and n2, where either magnesium or manganese bind | ||
| + | for catalysis. The n2 ion is involved in the phosphroyl transfer, while the n1 ion stabilizes an active GS and plays a role in binding glutamate <ref name="Eisenberg"/> . | ||
| + | <br> | ||
| + | <font size=4 face ="Arial">Involving Residues</font><br> | ||
| + | <font size=2>Most residues involved in enzymatic catalysis are located at the C domain but Asp50 is | ||
| + | contributed from the N domain of the other subunit <scene name='User:Grace_Natalie/Involving_residues_at_one_site/1'>(View of involving residues)</scene> . Both the N-terminus and C-terminus of each subunit are helical. | ||
| + | The N-terminal helix sits above the hexameric ring and is exposed to solvent <ref name="Eisenberg"/> . | ||
| + | The C-terminal hexlix (helical thong) is inserted into the hydrophobic hole in the subunit opposite hexameric ring | ||
| + | <ref name="Eisenberg"/> . The movement of Asp-50 aids in the formation of the ammonium binding site, and the movement of Arg-339 assist phosphoryl transfer and P<SUB>i</SUB> binding. <scene name='User:Grace_Natalie/Gln_synthetase_showing_asp50/1'>(Asp50 residue)</scene>.</font> | ||
| + | <center>[[Image:Untitled.JPG]]<br> | ||
| + | <font size=2>D. Eisneberg et al / Biochimica et Biophysica Acta 1477 (2000) 124</font></center> | ||
| + | <br><br> | ||
| + | <center><font size=4 face ="Arial">More Catalytic Residues</font><ref name="Liaw"/> | ||
| + | <table border=1> | ||
| + | <tr> | ||
| + | <td width=60><B>Residue</B></td> | ||
| + | <td width=370><B>Role in enzymatic mechanism</B></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width=60>Arg-321</td> | ||
| + | <td width=370>Coordinates the carboxylate of glutamate</td> | ||
| + | </tr><tr> | ||
| + | <td width=60>Glu-327</td> | ||
| + | <td width=370>Closes active site and shields intermediate from hydrolysis</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width=60>His-269</td> | ||
| + | <td width=370>Coordinates the n2 ion</td> | ||
| + | </tr><tr> | ||
| + | <td width=60>Glu-220</td> | ||
| + | <td width=370>Coordinates the n1 ion</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td width=60>Asp-50</td> | ||
| + | <td width=370>Increases the affinity for ammonium binding</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | </center> | ||
| + | <br><br> | ||
| + | <font size=4 face ="Arial">References</font> | ||
| + | <br> | ||
| + | <font size=2> <references/><br> | ||
| + | <br><br> | ||
| - | + | <br><br><br> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Background
|
Glutamine synthetase (GS) catalyzes the ATP-dependent condensation of ammonia and
glutamate to yield glutamine, ADP, and inorganic phosphate in the presence of divalent cations
[1] .
The reaction occurs in two steps with γ-glutamyl phosphate as an intermediate and is used by
bacteria to introduce reduced nitrogen into cellular metabolism. GS is a dodecamer formed from
two face-to-face hexameric rings of subunits, with 12 active sites formed between monomers
[2] .
Overall Mechanism
The first step is the formation of the activated intermediate γ-glutamyl phosphate.
The n2 ion coordinates the phosphate oxygens of ATP to allow phosphoryl transfer to the
γ-carboxylate group of glutatmate, yielding the intermediate [3] . The second step is the attack on the intermediate by ammonia
therefore releasing free phosphate to yield glutamine.
ATP binding site
Each active site of GS is described as a 'bifunnel in which ATP and glutamate bind at opposite ends.
The ATP binding site is referred to as the top of the bifunnel because it opens to the external 6-fold surface of GS (figure below) [3]. At the the joint of the are two cation binding sites, n1 and n2, where either magnesium or manganese bind
for catalysis. The n2 ion is involved in the phosphroyl transfer, while the n1 ion stabilizes an active GS and plays a role in binding glutamate [3] .
Involving Residues
Most residues involved in enzymatic catalysis are located at the C domain but Asp50 is
contributed from the N domain of the other subunit . Both the N-terminus and C-terminus of each subunit are helical.
The N-terminal helix sits above the hexameric ring and is exposed to solvent [3] .
The C-terminal hexlix (helical thong) is inserted into the hydrophobic hole in the subunit opposite hexameric ring
[3] . The movement of Asp-50 aids in the formation of the ammonium binding site, and the movement of Arg-339 assist phosphoryl transfer and Pi binding. .
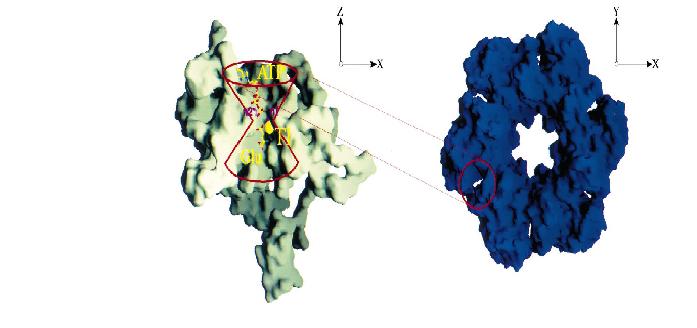
D. Eisneberg et al / Biochimica et Biophysica Acta 1477 (2000) 124
| Residue | Role in enzymatic mechanism |
| Arg-321 | Coordinates the carboxylate of glutamate |
| Glu-327 | Closes active site and shields intermediate from hydrolysis |
| His-269 | Coordinates the n2 ion |
| Glu-220 | Coordinates the n1 ion |
| Asp-50 | Increases the affinity for ammonium binding |
References
- ↑ 1.0 1.1 Liaw, S-H, et.al.,Discovery of the ammonium substrate site on glutamine synthetase, a third cation binding site Protein Sci. 1995 4: 2358-2365
- ↑ Gill, H & Eisenberg, D., Biochemistry 2001 40: 1903-1912
- ↑ 3.0 3.1 3.2 3.3 3.4 D. Eisneberg et al / Biochimica et Biophysica Acta 1477 (2000) 124
