This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
3ecp
From Proteopedia
(Difference between revisions)
| (9 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | {{Seed}} | ||
| - | [[Image:3ecp.png|left|200px]] | ||
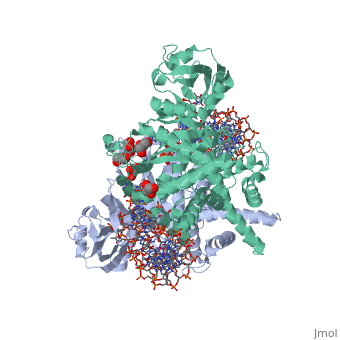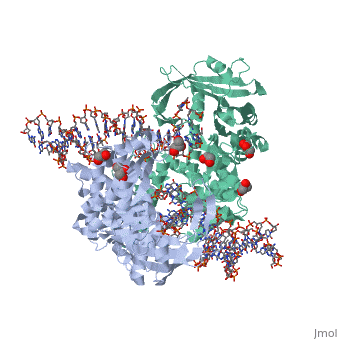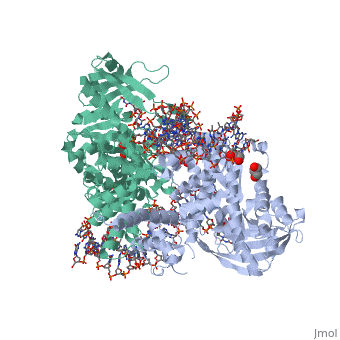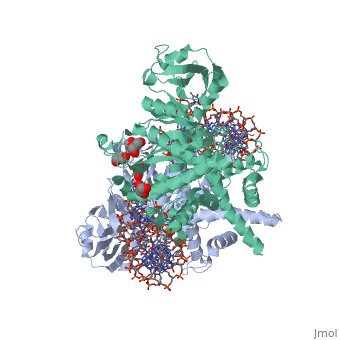
| - | < | + | ==Crystal Structure Of Tn5 Transposase Complexed With 5' Phosphorylated Transposon End DNA== |
| - | + | <StructureSection load='3ecp' size='340' side='right'caption='[[3ecp]], [[Resolution|resolution]] 2.50Å' scene=''> | |
| - | You may | + | == Structural highlights == |
| - | or the | + | <table><tr><td colspan='2'>[[3ecp]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3ECP OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3ECP FirstGlance]. <br> |
| - | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.5Å</td></tr> | |
| - | - | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=GOL:GLYCEROL'>GOL</scene></td></tr> |
| - | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3ecp FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3ecp OCA], [https://pdbe.org/3ecp PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3ecp RCSB], [https://www.ebi.ac.uk/pdbsum/3ecp PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3ecp ProSAT]</span></td></tr> | |
| + | </table> | ||
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/TN5P_ECOLX TN5P_ECOLX] Mediates transposition of transposon Tn5 by a 'cut and paste' mechanism. First, the monomeric transposase binds the 19 bp inverted DNA repeats flanking the transposon. Then, dimerization of the DNA-bound transposase creates a synaptic DNA complex. After nicking of the first DNA strand, excision of the transposon proceeds through a series of intermediates. The transposase then mediates the insertion of the transposon at a new site by strand transfer. The activity of the wild-type transposase is very low, and is further inhibited by dimerization with the transposase inhibitor (inh).<ref>PMID:6260374</ref> <ref>PMID:6291786</ref> <ref>PMID:6303899</ref> <ref>PMID:1310499</ref> <ref>PMID:8226636</ref> <ref>PMID:8871560</ref> <ref>PMID:11877443</ref> <ref>PMID:12367522</ref> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/ec/3ecp_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=3ecp ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | Bacterial DNA transposition is an important model system for studying DNA recombination events such as HIV-1 DNA integration and RAG-1-mediated V(D)J recombination. This communication focuses on the role of protein-phosphate contacts in manipulating DNA structure as a requirement for transposition catalysis. In particular, the participation of the nontransferred strand (NTS) 5' phosphate in Tn5 transposition strand transfer is analyzed. The 5' phosphate plays no direct catalytic role, nonetheless its presence stimulates strand transfer approximately 30-fold. X-ray crystallography indicates that transposase-DNA complexes formed with NTS 5' phosphorylated DNA have two properties that contrast with structures formed with complexes lacking the 5' phosphate or complexes generated from in-crystal hairpin cleavage. Transposase residues R210, Y319 and R322 of the (R)YREK motif coordinate the 5' phosphate rather than the subterminal NTS phosphate, and the 5' NTS end is moved away from the 3' transferred strand end. Mutation R210A impairs the 5' phosphate stimulation. It is posited that DNA phosphate coordination by R210, Y319 and R322 results in movement of the 5' NTS DNA away from the 3'-end thus allowing efficient target DNA binding. It is likely that this role for the newly identified RYR triad is utilized by other transposase-related proteins. | ||
| - | + | Phosphate coordination and movement of DNA in the Tn5 synaptic complex: role of the (R)YREK motif.,Klenchin VA, Czyz A, Goryshin IY, Gradman R, Lovell S, Rayment I, Reznikoff WS Nucleic Acids Res. 2008 Oct;36(18):5855-62. Epub 2008 Sep 12. PMID:18790806<ref>PMID:18790806</ref> | |
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
| + | <div class="pdbe-citations 3ecp" style="background-color:#fffaf0;"></div> | ||
| - | + | ==See Also== | |
| - | + | *[[Transposase|Transposase]] | |
| - | + | *[[Transposase 3D structures|Transposase 3D structures]] | |
| - | + | == References == | |
| - | + | <references/> | |
| - | + | __TOC__ | |
| - | == | + | </StructureSection> |
| - | + | ||
| - | + | ||
| - | == | + | |
| - | + | ||
[[Category: Escherichia coli]] | [[Category: Escherichia coli]] | ||
| - | [[Category: | + | [[Category: Large Structures]] |
| - | [[Category: | + | [[Category: Klenchin VA]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Crystal Structure Of Tn5 Transposase Complexed With 5' Phosphorylated Transposon End DNA
| |||||||||||