Anderson Sandbox
From Proteopedia
m |
|||
| (3 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | ===A SMART Team Modeling Project from the CBM=== | ||
| - | ---- | ||
| - | |||
| - | ====This is a Test Series of scenes for <scene name='Anderson_Sandbox/Orig/1'>Lac-Y Permease</scene>==== | ||
| - | |||
| - | To the right is an image representation of Lac-Y permese of E-coli. The Ligand is shown <scene name='Anderson_Sandbox/Ligand/3'>here</scene>, the Sidechains Participating in substrate binding are <scene name='Anderson_Sandbox/Sbu_binding_res/2'>here</scene>, and residues participating in proton translocation are here. | ||
| - | |||
| - | ====Function of Lactose Permease==== | ||
| - | [[Image:(image info)|thumb|(Caption)]] | ||
| - | |||
| - | |||
| - | Lactose Permease is a transmembrane protein that facilitates the passage of lactose across the phospholipid bi-layer of the cell membrane. The transport mechanism used is an active co-transport that uses the inwardly directed H+ electrochemical gradient as its driving force. As a result, the lactose is accompanied from the periplasm to the cytoplasm of the cell by an H+ proton.<ref name="A. Green, et. al.">[http://www.jbc.org/cgi/content/full/275/30/23240]</ref> | ||
| - | |||
| - | Lactose is a disaccharide carbohydrate found primarily in mammalian milk. It is a disaccharide composed of the monosaccharides glucose and galactose. When lactose is ingested, it is brought into cells in the digestive system by the protein Lactose Permease. Here it is broken down into its monosaccharide subunits by the enzyme lactase so it may be used in the process of cellular respiration.<ref name="G. Gidwani">[http://www.faqs.org/nutrition/Kwa-Men/Lactose-Intolerance.html]</ref> | ||
| - | |||
| - | <applet load='1pv7' size='350' frame='true' align='right' caption='Image of Lac-y Permease' /> | ||
| - | |||
| - | Highlighting the <scene name='Anderson_Sandbox/All_sulfurs/1'>Sulfur</scene> of the lactose permease protein. | ||
| - | |||
| - | ---- | ||
| - | Replace the PDB id after the STRUCTURE_ and after PDB= to load | ||
| - | and display another structure. | ||
| - | |||
| - | {{STRUCTURE_ | PDB= | SCENE= }} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
---- | ---- | ||
| Line 52: | Line 23: | ||
---- | ---- | ||
| + | ==Center for Biomolecular Modeling Physical Models== | ||
| + | The MSOE Center for Biomolecular Modeling has created various physical models of Hemoglobin. Posted is a video displaying one such model and a discussion of some of the properites of Hemoglobin it exhibits. For more information about the CBM and the models we create, visit our site at [http://cbm.msoe.edu http://cbm.msoe.edu]. | ||
| + | |||
| + | |||
| + | ---- | ||
| + | |||
==Different CBM Slogans== | ==Different CBM Slogans== | ||
Current revision
What is a SMART Team?
S- Students M- Modeling A R- Research T- Topic
This is a program currently run by the Center for Biomolecular Modeling headed by Shannon Colton. We at the CBM do not do any of the research it's all the students that research and study what it is that their molecule does. These students are any where from Jr.High students to Seniors graduating from High School. Our job at the CBM is to put them in contact with a researcher that is interested in participating in the program and to help in making a physical model for the students to use when demonstrating what it is their molecule does. www.rpc.msoe.edu/cbm/smartteams/
Why Should I join a SMART team?
- It's a great way to be introduced to modern research
- Looks good on a College application
- Good way to meet new people that are interested in the same things you are
- Most Importantly it's FUN!!
Center for Biomolecular Modeling Physical Models
The MSOE Center for Biomolecular Modeling has created various physical models of Hemoglobin. Posted is a video displaying one such model and a discussion of some of the properites of Hemoglobin it exhibits. For more information about the CBM and the models we create, visit our site at http://cbm.msoe.edu.
Different CBM Slogans
The physical models shown on this page were designed and built by the MSOE Center for BioMolecular Modeling. For more information about physical protein modeling, visit the CBM web site at www.rpc.msoe.edu/cbm .
The physical models displayed on this webpage have been designed and build by the MSOE Center for BioMolecular Modeling. To learn more about the CBM and see more examples of protein models, please visit the CBM site at www.rpc.msoe.edu/cbm.
The physical models displayed here are examples of models designed and built by the MSOE Center for BioMolecular Modeling. To see more modeling possibilities and learn more about the CBM, please visit our site at www.rpc.msoe.edu/cbm.
Physical Models
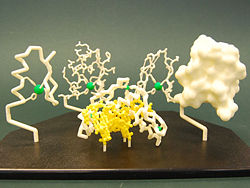
The model shown in Model Image 1 to the left is an example of a physical model of the zinc finger protien. In this representation, the Alpha Helix is shown in Red, the Beta Sheets are displayed in Yellow, and the Residues are represented in CPK color format (O=red, N=blue, C=gray, S=yellow). The Zinc Atom in this structure is colored Red.
The model in Model Image 2 shown to the right is an example of a 'mini-toober' model. The side chains are again displayed in CPK color format. The zinc atom in this example is displayed in green.
MSOE Center for BioMolecular Modeling
The physical models shown on this page were designed and built by the MSOE Center for BioMolecular Modeling. For more information about physical protein modeling, visit the CBM web site at www.rpc.msoe.edu/cbm .
MSOE Center for BioMolecular Modeling

The physical models shown on this page were designed and built by the MSOE Center for BioMolecular Modeling. For more information about physical protein modeling, visit the CBM web site at www.rpc.msoe.edu/cbm .
MSOE Center for BioMolecular Modeling

The physical models shown on this page were designed and built by the MSOE Center for BioMolecular Modeling. For more information about physical protein modeling, visit the CBM web site at www.rpc.msoe.edu/cbm .
MSOE Center for BioMolecular Modeling

The physical models shown on this page were designed and built by the MSOE Center for BioMolecular Modeling. For more information about physical protein modeling, visit the CBM web site at www.rpc.msoe.edu/cbm .
MSOE Center for BioMolecular Modeling

The physical models shown on this page were designed and built by the MSOE Center for BioMolecular Modeling. For more information about physical protein modeling, visit the CBM web site at www.rpc.msoe.edu/cbm .
Physical Models
The model shown in Model Image 1 to the left is an example of a physical model of the zinc finger protien. In this representation, the Alpha Helix is shown in Red, the Beta Sheets are displayed in Yellow, and the Residues are represented in CPK color format (O=red, N=blue, C=gray, S=yellow). The Zinc Atom in this structure is colored Red.
The model in Model Image 2, shown in the center, is an example of a 'mini-toober' model. The side chains are again displayed in CPK color format. The zinc atom in this example is displayed in green.
The models in Model Image 3 are a variety of different representations for the zinc finger molecule. These representations include backbone, wireframe, and spacefill model types.
MSOE Center for BioMolecular Modeling
The physical models shown on this page were designed and built by the MSOE Center for BioMolecular Modeling. For more information about physical protein modeling, visit the CBM web site at www.rpc.msoe.edu/cbm .