Sandbox 4
From Proteopedia
| (49 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | [[Image:1emaBiomics4.gif|350 px]] | |
| - | [[Image: | + | |
| - | <!-- | ||
| - | The line below this paragraph, containing "STRUCTURE_1tsj", creates the "Structure Box" on the page. | ||
| - | You may change the PDB parameter (which sets the PDB file loaded into the applet) | ||
| - | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | ||
| - | or leave the SCENE parameter empty for the default display. | ||
| - | --> | ||
| - | + | Green fluorescent protein ('''GFP'''), originally isolated from the jellyfish Aequorea victoria (PDB entry [[1ema]]), fluorsceses green (509nm) when exposed to blue light (395nm and 475nm). It is one of the most important proteins used in biological research because it can be used to tag otherwise invisible gene products of interest and thus observe their existence, location and movement. | |
| - | PDB entry 1tsj refers to a hypothetical protein of 139 residues which is a predicted dimer<ref> Henrick, K., and Thornton, J.M. (1998). PQS: a protein quaternary structure file server. Trends Biochem. Sci. 23, 358- 361.</ref> and predicted as a cytoplasmic protein.<ref> Bhasin, M., Garg, A. and Raghava, GPS (2005) PSLpred: prediction of subcellular localization of bacterial proteins. Bioinformatics. 21, 2522-2524. | ||
| - | Gardy JL, Spencer C, Wang K, Ester M, Tusn?dy GE, Simon I, Hua S, deFays K, Lambert C, Nakai K, Brinkman FS. (2003) PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 31, 3613-3617. | ||
| - | Z. Lu, D. Szafron, R. Greiner, P. Lu, D.S. Wishart, B. Poulin, J. Anvik, C. Macdonell, and R. Eisner. (2003). Predicting Subcellular Localization of Proteins using Machine-Learned Classifiers. Bioinformatics. 20, 547-556. | ||
| - | Hua S, Sun Z. (2001). Support vector machine approach for protein subcellular localization prediction. Bioinformatics. 17, 721-728.</ref> | ||
| - | + | == Exploring the Structure == | |
| + | <applet load='1ema' size='300' frame='true' align='right' caption='Insert caption here' /> | ||
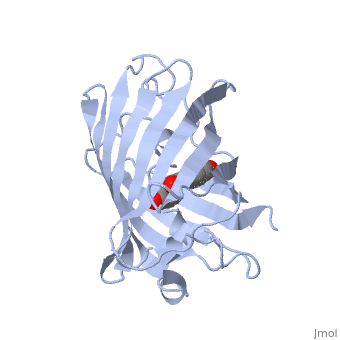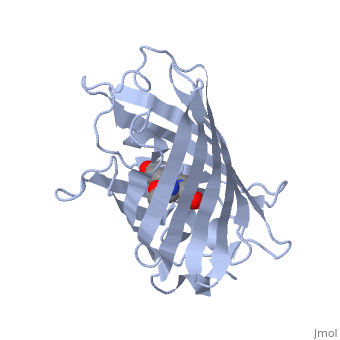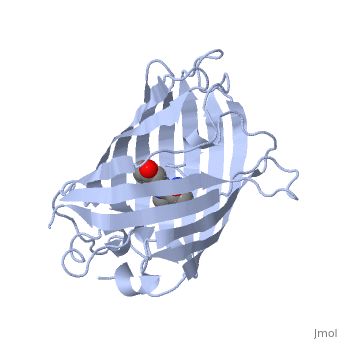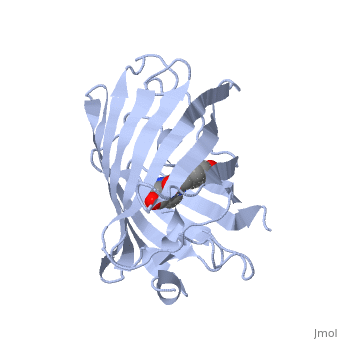
| - | + | GFP is a beta barrel protein with 11 beta sheets. It is a 26.9kDa protein made up of 238 amino acids. The | |
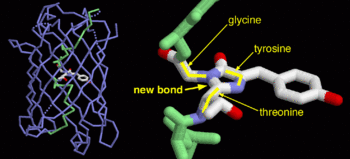
| - | + | <scene name='Sandbox_4/Green_fp/1'>chromophore</scene>, responsible for the fluorescent properties of the protein, is buried inside the beta barrel as part of the central alpha helix passing through the barrel. The chromophore forms via spontaneous cyclization and oxidation of three residues in the central alpha helix: -Thr65 (or Ser65)-Tyr66-Gly67. This cyclization and oxidation creates the chromophore's five-membered ring via a new bond between the threonine and the glycine residues.<ref>PMID:8703075</ref> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==References== | ==References== | ||
<references/> | <references/> | ||
| - | |||
| - | [[Category: Staphylococcus aureus subsp. aureus]] | ||
| - | [[Category: Burley, S K.]] | ||
| - | [[Category: Gorman, J.]] | ||
| - | [[Category: Min, T.]] | ||
| - | [[Category: NYSGXRC, New York Structural GenomiX Research Consortium.]] | ||
| - | [[Category: Shapiro, L.]] | ||
| - | [[Category: Conserved hypothetical protein]] | ||
| - | [[Category: Crystal structure]] | ||
| - | [[Category: New york structural genomics consortium]] | ||
| - | [[Category: New york structural genomix research consortium]] | ||
| - | [[Category: Nysgxrc]] | ||
| - | [[Category: Protein structure initiative]] | ||
| - | [[Category: Psi]] | ||
| - | [[Category: Structural genomic]] | ||
Current revision
Green fluorescent protein (GFP), originally isolated from the jellyfish Aequorea victoria (PDB entry 1ema), fluorsceses green (509nm) when exposed to blue light (395nm and 475nm). It is one of the most important proteins used in biological research because it can be used to tag otherwise invisible gene products of interest and thus observe their existence, location and movement.
Exploring the Structure
|
GFP is a beta barrel protein with 11 beta sheets. It is a 26.9kDa protein made up of 238 amino acids. The , responsible for the fluorescent properties of the protein, is buried inside the beta barrel as part of the central alpha helix passing through the barrel. The chromophore forms via spontaneous cyclization and oxidation of three residues in the central alpha helix: -Thr65 (or Ser65)-Tyr66-Gly67. This cyclization and oxidation creates the chromophore's five-membered ring via a new bond between the threonine and the glycine residues.[1]