This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox 19
From Proteopedia
(Difference between revisions)
| (2 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | ==This is a placeholder== | |
| + | This is a placeholder text to help you get started in | ||
| + | placing a Jmol applet on your page. At any time, click | ||
| + | "Show Preview" at the bottom of this page to see how it goes. | ||
| + | Replace the PDB id (use lowercase!) after the STRUCTURE_ and after PDB= to load | ||
| + | and display another structure. | ||
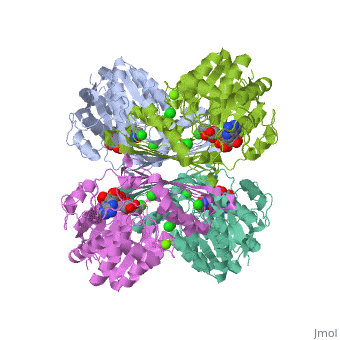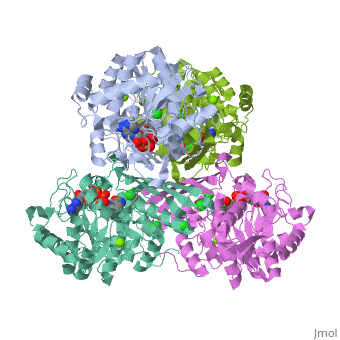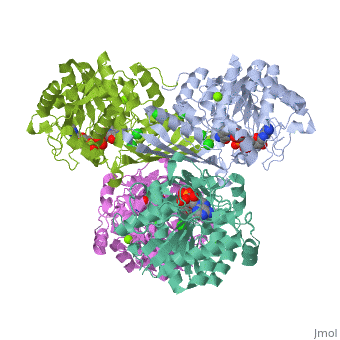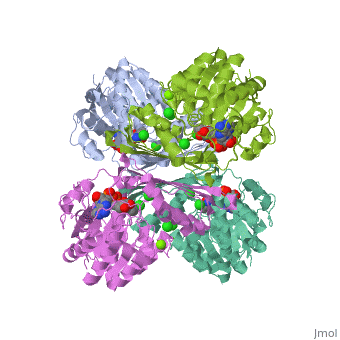
| - | == | + | {{STRUCTURE_3cin | PDB=3cin | SCENE= }} |
| - | + | ||
| - | + | <scene name='Sandbox_19/Emre/1'>TextToBeDisplayed</scene> | |
Current revision
This is a placeholder
This is a placeholder text to help you get started in placing a Jmol applet on your page. At any time, click "Show Preview" at the bottom of this page to see how it goes.
Replace the PDB id (use lowercase!) after the STRUCTURE_ and after PDB= to load and display another structure.
| |||||||||
| 3cin, resolution 1.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | TM1419, TM_1419 (Thermotoga maritima MSB8) | ||||||||
| Activity: | Inositol-3-phosphate synthase, with EC number 5.5.1.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||