User:Ramiro Barrantes/FpgNeiRepair
From Proteopedia
(→Latent Structural Characters (LSCs)) |
|||
| (194 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | + | __NOTOC__ | |
| - | == | + | = The FpgNei Protein Family = |
| - | + | ||
| - | + | [[#Background on DNA Repair|1. Background on DNA Repair]] <br> | |
| + | [[#Background on Fpg/Nei|2. Background on Fpg/Nei]]<br> | ||
| + | [[#Overall Function and Structure|2.1 Overall Function and Structure]]<br> | ||
| + | [[#Latent Structural Characters (LSCs)|3. Latent Structural Characters Summary]]<br> | ||
| + | [[#References|5. References]] | ||
| - | == Background on | + | ==Background on DNA Repair== |
| - | + | The genome of any living organism is continuously affected by exogenous and endogenous agents, such as ultraviolet light, ionizing radiation, different chemicals and the cell's own metabolites (such as reactive oxygen). Therefore, different systems have evolved to repair these damages, with some of these systems shared throughout all lifeforms. Therefore, the proper functioning of DNA repair is critical for survival. There are six pathways of DNA repair (reviewed in Friedberg et al), which include base-excision repair. The latter's distinguishing feature is that it removes lesions as single bases, as opposed to dNMPs or short oligonucleotides like other systems. <ref>PMID:18259689</ref> | |
| - | + | Base excision repair's signature enzyme are the DNA glycosylases. These enzymes work by recognizing a damaged base, and then hydrolizing the N-glycosidic bond of the damaged deoxynucleoside and thus removing a single damaged base from DNA. The subsequent steps of the pathway (strand incision, gap-filling and ligation) are done by other enzymes. <ref>PMID: 18259689</ref><ref>PMID:19153658</ref> | |
| - | + | ||
| - | + | ||
| - | + | ==Background on Fpg Nei== | |
| - | + | ==Overall Function and Structure== | |
| - | + | {| valign="top" | |
| - | + | | valign="top"|(For FPG, the structure used was [[1r2y]] and for Nei [[1k3w]]) Members of this family have <scene name='User:Ramiro_Barrantes/Workbench/Gstfpglabeleddomains/4' target="function"> two domains (blue and orange) connected by a hinge region (purple). DNA (gray) binds in a cleft between the domains. </scene>. When enzyme binds to DNA, the damaged base | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <scene name='User:Ramiro_Barrantes/Workbench/Evertedbaseonbstfpg/1' target="function"> is everted</scene>, with <scene name='User:Ramiro_Barrantes/Workbench/Gstfpgintercalationloop/2' target="function"> several residues intercalating in its place</scene>. This superfamily is also characterized by containing a <scene name='User:Ramiro_Barrantes/Workbench/Gstfpgh2th/2' target="function">helix-two-turn-helix motif (H2TH, in purple) which contacts DNA via Asn174 (in yellow)</scene>; as well as a | ||
| + | <scene name='User:Ramiro_Barrantes/Workbench/Gstfpgzincfinger/5' target="function"> zinc finger, which contacts DNA via Arg264 </scene>. Both the H2TH motif and the Zinc finger, as well as other residues | ||
| + | <scene name='User:Ramiro_Barrantes/Workbench/Gstfpgdamagestabilizers/1' target="function"> stabilize the damaged base </scene>. <ref>PMID:11912217</ref>. Catalysis is believed to be mediated by <scene name='User:Ramiro_Barrantes/Workbench/Gstfpgcatalyticresidues/3' target="function">P2, E3, E6 and R264</scene>. For information on the mechanism please consult <ref>PMID:16243784</ref><ref>PMID:11847126</ref><ref>PMID:15249553</ref><ref>PMID:11912217</ref><ref>PMID:10921868</ref>. | ||
| + | |||
| + | Similarly In EcoNei (structure [[1k3w]], <scene name='User:Ramiro_Barrantes/Workbench/Econeiorientationandkinking/4' target="function">E2,K52,N168 and R252 provide orientation and kinking to the DNA,</scene>, and <scene name='User:Ramiro_Barrantes/Workbench/Econeistabilizingsites/2' target="function">P1, E5 and R212 and others stabilize the damaged base </scene>(Thymine Glycol in the case of Nei). <ref>PMID:11847126</ref>. Some of these amino acids are stabilized by [a <scene name='User:Ramiro_Barrantes/Workbench/Econeizincfinger/1' target="function"> Zinc Finger </scene> (although a zincless finger motif is present in some of these subfamilies <ref>PMID:15232006</ref>). Analogously to Fpg, <scene name='User:Ramiro_Barrantes/Workbench/Econeiintercalationloop/4'>''E. coli Nei'' residues insert into the vacated spot left by the "flipped" base</scene>), Note that the last two elements discussed, the zinc vs. zincless finger, and the two kinds of intercalation loops, are examples of coevolving functional clusters, groups of amino acids that perform a function, and that might be unnecessary or compensated for within the other subfamilies. We developed a novel method for identifying these clusters and have applied it to bring insight into the structure, function and evolution FpgNei family. | ||
| + | | <applet load='1K3W' size='500' frame='true' align='left' caption='See structure column for structure reference' name="function" scene='User:Ramiro_Barrantes/Workbench/Gstfpglabeleddomains/3' /> | ||
| + | |- | ||
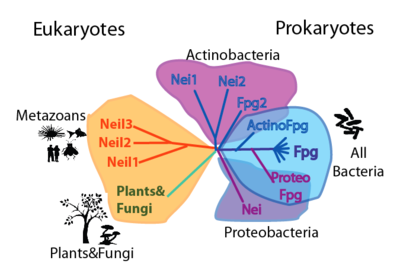
| + | | valign="top"|===Phylogeny=== [[Image:Picture 9.png|thumb|400px|left|Cartoon phylogenetic tree of the FpgNei protein family. Note that this phylogeny can be appreciated in two levels: by the distribution and number of FpgNei subfamilies in different organisms; and by the kinds of damages that can be repaired (see table on the right)]] | ||
| + | | valign="top"|===Table of preferred substrates=== | ||
{| class="wikitable" style="text-align:center" | {| class="wikitable" style="text-align:center" | ||
|- | |- | ||
| - | ! | + | ! Clade !! Function !! References |
|- | |- | ||
| - | | | + | | Fpg || 8oxoG, Fapy-A, Fapy-G,Me-Fapy-G,Sp,Gh || <ref>PMID:11101315</ref><ref>PMID:11848931</ref> |
| - | | < | + | |
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | | EcoNei || Oxidized pyrimidines || <ref>PMID:9405426</ref> |
| - | | < | + | |
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | | Actinomycetes Nei1 || DHU || <ref>PMID:18457574</ref> |
| - | | < | + | |
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | | Actinomycetes Nei2 || ? || |
| - | | | + | |
| - | | | + | |
| - | | | + | |
|- | |- | ||
| - | | | + | | Fpg2 || ? || |
| - | | | + | |
| - | | | + | |
| - | | | + | |
|- | |- | ||
| - | | | + | | Plant&Fungi || Sp,Gh || <ref>PMID:19217358</ref> |
| - | | | + | |
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | | Neil1 || Sp,Gh,on double&single strand DNA, Both stereoisomers of Tg|| <ref>PMID:15533836</ref><ref>PMID:12713815</ref> |
| - | | < | + | |- |
| - | | | + | | Neil2 || Sp,Gh,on double&single stranded DNA || <ref>PMID:15533836</ref> |
| - | | | + | |- |
| + | | Neil3 || not clear || <ref>PMID:19170771</ref> | ||
| + | |} | ||
|} | |} | ||
| - | + | ===Mechanisms=== | |
| - | == | + | Several authors have suggested mechanisms for these enzymes, please see references for more information <ref>PMID:11847126</ref><ref>PMID:10921868</ref><ref>PMID:16243784</ref><ref>PMID:11912217</ref>. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | === | + | ===Solved Structures=== |
| - | + | Homologous structures have been solved, including Fpg protein from Lactococcus Lactis ([[1pjj]])<ref>PMID: 16243784</ref>, Bacillus Stereothermophilus ([[1r2y]])<ref>PMID:14525999</ref>, Thermos Thermophilus ([[1ee8]])<ref>PMID:10921868</ref> and Escherichia Coli([[1k82]])<ref>PMID:11912217</ref> and Nei from Escherichia Coli ([[1k3w]])<ref>PMID:11847126</ref>, as well as human Neil1 ([[1tdh]])<ref>PMID:19625256</ref>and mimivirus Nei ([[3a42]])<ref>PMID:17627905</ref>. The overall structure is similar, and some of the damages include 8-oxoguanine and fapyG ([[1xc8]])<ref>PMID:15249553</ref>. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | ==Latent Structural Characters (LSCs)== |
| - | [[Image: | + | {| |
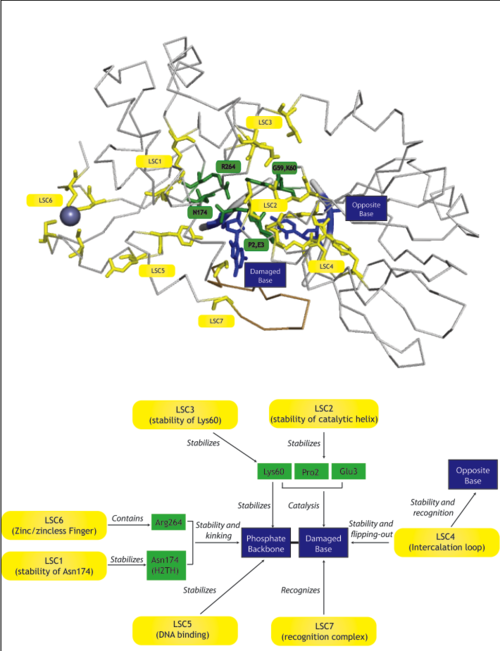
| + | | [[Image:LSCDiagram.png|thumb|500px|right|Diagram with the LSCs (yellow), their suggested role (arrows) and the amino acids or features they affect (green and blue)]] We define a latent structural character (LSC) as neighboring amino acids which have changed in rate or constraint for a given clade or set of clades. For example, the cysteines in the zinc finger are all conserved in the fpg1, fpg2, actinobacterial and eukaryotic clades, but have a higher rate in plants and neil1, as the latter have a zinc-less finger (see [http://proteopedia.org/wiki/index.php?title=User:Ramiro_Barrantes/Workbench/LSC6 here] for more information). We developed a novel method to find these groups of amino acids, discovering previously unknown groups of amino acids which are functionally important in some of these subfamilies. | ||
| + | The table below has some of our main findings, there are two variants for each LSC in case that for one LSC there is a compensating cluster in the other subfamilies. If you click on one of them you will see the result in the structure, and you can go to the explanation and the sequence distribution below. <br> [http://xpdb.nist.gov/hivsdb/jmol/jmol_help.html Click here for Jmol tutorial].| | ||
| + | |} | ||
| - | == | + | {| - style="font-size:18px;align=center" |
| - | + | | <applet load='1R2Y' size='500' frame='true' align='left' name="fpg" scene='User:Ramiro_Barrantes/Workbench/Gstfpg/5'/> | |
| - | + | | <applet load='1K3W' size='500' frame='true' align='right' name="ecoNei" scene='User:Ramiro_Barrantes/Workbench/Econeiwater/1' /> | |
| + | |} | ||
| - | + | {| class="wikitable" style="text-align:center" | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | {| class="wikitable" style="text-align:center" | + | |
| - | + | ||
|- | |- | ||
| - | + | ! Functional Cluster !! State 1 !! State 2 !! Fpg1 !! Fpg2 !! Plant !! Neil1 !! Neil2 !! Neil3 !! Proteo !! Actino1 !! Actino2 !! MimiVirus | |
|- | |- | ||
| - | | <scene name='User:Ramiro_Barrantes/Workbench/ | + | | Support for perfectly conserved residue |
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Lys160asp178/13' target="fpg">Lys160 and Asp178</scene> | ||
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Econeiarginine171/10' target="ecoNei">Arg171</scene> | ||
| + | | 1 || 1 || 1 || 0 || 0 || 0 || 0 || 0 || 0 || 0 | ||
|- | |- | ||
| - | | <scene name='User:Ramiro_Barrantes/Workbench/ | + | | Stability of catalytic helix |
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Gstfpgglu8arg57leu4triad/8' target="fpg">Leu4,Glu8,Arg57</scene> | ||
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Econeisupporttriadcounterpart/5' target="ecoNei">Different</scene> | ||
| + | | 1 || 1 || 1 || - || - || - || - || - || - || - | ||
|- | |- | ||
| - | | <scene name='User:Ramiro_Barrantes/Workbench/ | + | | Neil1-specific: Stability of Lys60 |
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Gstfpgneil1covariate/5' target="fpg">E137,R58,G135,L134</scene> | ||
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Neil1lys60support/3' target="ecoNei">Covariation with Asn172</scene> | ||
| + | | 1 || 1 || 1|| 0 || 1 || 1 || 1 || 1 || 1 || 1 | ||
|- | |- | ||
| - | | <scene name='User:Ramiro_Barrantes/Workbench/ | + | | Intercalation loop |
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Fpgintercalationloop/3' target="fpg">D110,F108,R113,R112,F114</scene> | ||
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Econeiintercalationloop/4' target="ecoNei">Gln76,Met77,Tyr78</scene> | ||
| + | | 1 || 1 || 1 || - || - || - || - || - || - || 1 | ||
|- | |- | ||
| - | | <scene name='User:Ramiro_Barrantes/Workbench/ | + | | PlantFungi-specific: R254 DNA binding |
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Gstfpgtyr243/4' target="fpg">Y242,G243,R244</scene> | ||
| + | | different in plants | ||
| + | | 1 || 1 || 0 || 1 || 1 || 1 || 2 || 1 || 1 || 1 | ||
|- | |- | ||
| - | | <scene name='User:Ramiro_Barrantes/Workbench/ | + | | Zinc/zincless finger |
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Gstfpgzincfinger/6' target="fpg">Zinc finger</scene> | ||
| + | | <scene name='User:Ramiro_Barrantes/Workbench/Neil1zinclessfinger/5' target="ecoNei">Neil1 zincless finger</scene> | ||
| + | | 1 || 1 || - || - || 1 || 1 || 1 || 1 || 1 || - | ||
|- | |- | ||
| - | | | + | | Recognition complex |
| - | + | | <scene name='User:Ramiro_Barrantes/Workbench/Gstfpgrecognitioncomplex/1' target="fpg">Loop from Gly218 to Gly233</scene> | |
| - | | <scene name='User:Ramiro_Barrantes/Workbench/ | + | | <scene name='User:Ramiro_Barrantes/Workbench/LSCs/Neil1helixalpha10beta9/1' target="ecoNei">Helix in Neil1</scene> |
| - | + | | 1 || - || - || - || - || - || - || - || - || - | |
| - | | <scene name='User:Ramiro_Barrantes/Workbench/ | + | |
| - | + | ||
| - | + | ||
| - | | | + | |
| - | + | ||
| - | |- | + | |
| - | | | + | |
| - | |- | + | |
| - | | | + | |
| - | |- | + | |
| - | | | + | |
| - | |- | + | |
| - | | | + | |
| - | |- | + | |
| - | | | + | |
| - | |- | + | |
| - | | | + | |
| - | |- | + | |
| - | | | + | |
| - | |- | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| + | |||
| + | == Evolution == | ||
| + | The FpgNei evolution has not been easy to resolve <ref>PMID:15232006</ref>, especially in the deeper branches. Assuming that functional clusters evolve more slowly than individual residues, we can use this as phylogenetic characters to 1) draw the most parsimonious evolution of the superfamily as dictated by these functional clusters 2) examine how these clusters have evolved and how this might have influenced the evolution of FpgNei. | ||
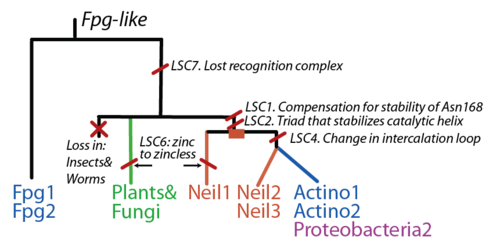
| + | [[Image:Scenario1.png|thumb|500px|right|Gene evolution of the Fpg/Nei protein family. The root is placed in the Fpg branch given its broad distribution among bacteria.)]] | ||
==References== | ==References== | ||
<references /> | <references /> | ||
Current revision
The FpgNei Protein Family
1. Background on DNA Repair
2. Background on Fpg/Nei
2.1 Overall Function and Structure
3. Latent Structural Characters Summary
5. References
Background on DNA Repair
The genome of any living organism is continuously affected by exogenous and endogenous agents, such as ultraviolet light, ionizing radiation, different chemicals and the cell's own metabolites (such as reactive oxygen). Therefore, different systems have evolved to repair these damages, with some of these systems shared throughout all lifeforms. Therefore, the proper functioning of DNA repair is critical for survival. There are six pathways of DNA repair (reviewed in Friedberg et al), which include base-excision repair. The latter's distinguishing feature is that it removes lesions as single bases, as opposed to dNMPs or short oligonucleotides like other systems. [1]
Base excision repair's signature enzyme are the DNA glycosylases. These enzymes work by recognizing a damaged base, and then hydrolizing the N-glycosidic bond of the damaged deoxynucleoside and thus removing a single damaged base from DNA. The subsequent steps of the pathway (strand incision, gap-filling and ligation) are done by other enzymes. [2][3]
Background on Fpg Nei
Overall Function and Structure
| (For FPG, the structure used was 1r2y and for Nei 1k3w) Members of this family have . When enzyme binds to DNA, the damaged base
, with . This superfamily is also characterized by containing a ; as well as a . Both the H2TH motif and the Zinc finger, as well as other residues . [4]. Catalysis is believed to be mediated by . For information on the mechanism please consult [5][6][7][8][9]. Similarly In EcoNei (structure 1k3w, , and (Thymine Glycol in the case of Nei). [10]. Some of these amino acids are stabilized by [a (although a zincless finger motif is present in some of these subfamilies [11]). Analogously to Fpg, ), Note that the last two elements discussed, the zinc vs. zincless finger, and the two kinds of intercalation loops, are examples of coevolving functional clusters, groups of amino acids that perform a function, and that might be unnecessary or compensated for within the other subfamilies. We developed a novel method for identifying these clusters and have applied it to bring insight into the structure, function and evolution FpgNei family. |
| ||||||||||||||||||||||||||||||
| ===Phylogeny=== | ===Table of preferred substrates===
|
Mechanisms
Several authors have suggested mechanisms for these enzymes, please see references for more information [21][22][23][24].
Solved Structures
Homologous structures have been solved, including Fpg protein from Lactococcus Lactis (1pjj)[25], Bacillus Stereothermophilus (1r2y)[26], Thermos Thermophilus (1ee8)[27] and Escherichia Coli(1k82)[28] and Nei from Escherichia Coli (1k3w)[29], as well as human Neil1 (1tdh)[30]and mimivirus Nei (3a42)[31]. The overall structure is similar, and some of the damages include 8-oxoguanine and fapyG (1xc8)[32].
Latent Structural Characters (LSCs)
| We define a latent structural character (LSC) as neighboring amino acids which have changed in rate or constraint for a given clade or set of clades. For example, the cysteines in the zinc finger are all conserved in the fpg1, fpg2, actinobacterial and eukaryotic clades, but have a higher rate in plants and neil1, as the latter have a zinc-less finger (see here for more information). We developed a novel method to find these groups of amino acids, discovering previously unknown groups of amino acids which are functionally important in some of these subfamilies.
The table below has some of our main findings, there are two variants for each LSC in case that for one LSC there is a compensating cluster in the other subfamilies. If you click on one of them you will see the result in the structure, and you can go to the explanation and the sequence distribution below. |
|
|
| Functional Cluster | State 1 | State 2 | Fpg1 | Fpg2 | Plant | Neil1 | Neil2 | Neil3 | Proteo | Actino1 | Actino2 | MimiVirus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Support for perfectly conserved residue | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Stability of catalytic helix | 1 | 1 | 1 | - | - | - | - | - | - | - | ||
| Neil1-specific: Stability of Lys60 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| Intercalation loop | 1 | 1 | 1 | - | - | - | - | - | - | 1 | ||
| PlantFungi-specific: R254 DNA binding | different in plants | 1 | 1 | 0 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | |
| Zinc/zincless finger | 1 | 1 | - | - | 1 | 1 | 1 | 1 | 1 | - | ||
| Recognition complex | 1 | - | - | - | - | - | - | - | - | - |
Evolution
The FpgNei evolution has not been easy to resolve [33], especially in the deeper branches. Assuming that functional clusters evolve more slowly than individual residues, we can use this as phylogenetic characters to 1) draw the most parsimonious evolution of the superfamily as dictated by these functional clusters 2) examine how these clusters have evolved and how this might have influenced the evolution of FpgNei.
References
- ↑ Zharkov DO. Base excision DNA repair. Cell Mol Life Sci. 2008 May;65(10):1544-65. PMID:18259689 doi:10.1007/s00018-008-7543-2
- ↑ Zharkov DO. Base excision DNA repair. Cell Mol Life Sci. 2008 May;65(10):1544-65. PMID:18259689 doi:10.1007/s00018-008-7543-2
- ↑ Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009 Mar;66(6):981-93. PMID:19153658 doi:10.1007/s00018-009-8736-z
- ↑ Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002 May 31;277(22):19811-6. Epub 2002 Mar 23. PMID:11912217 doi:http://dx.doi.org/10.1074/jbc.M202058200
- ↑ Pereira de Jesus K, Serre L, Zelwer C, Castaing B. Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 2005 Oct 20;33(18):5936-44. Print 2005. PMID:16243784 doi:http://dx.doi.org/33/18/5936
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Coste F, Ober M, Carell T, Boiteux S, Zelwer C, Castaing B. Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase. J Biol Chem. 2004 Oct 15;279(42):44074-83. Epub 2004 Jul 10. PMID:15249553 doi:10.1074/jbc.M405928200
- ↑ Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002 May 31;277(22):19811-6. Epub 2002 Mar 23. PMID:11912217 doi:http://dx.doi.org/10.1074/jbc.M202058200
- ↑ Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000 Aug 1;19(15):3857-69. PMID:10921868 doi:http://dx.doi.org/10.1093/emboj/19.15.3857
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci U S A. 2004 Jul 13;101(28):10284-9. Epub 2004 Jul 1. PMID:15232006 doi:10.1073/pnas.0402051101
- ↑ Leipold MD, Muller JG, Burrows CJ, David SS. Removal of hydantoin products of 8-oxoguanine oxidation by the Escherichia coli DNA repair enzyme, FPG. Biochemistry. 2000 Dec 5;39(48):14984-92. PMID:11101315
- ↑ David SS, Williams SD. Chemistry of Glycosylases and Endonucleases Involved in Base-Excision Repair. Chem Rev. 1998 May 7;98(3):1221-1262. PMID:11848931
- ↑ Jiang D, Hatahet Z, Melamede RJ, Kow YW, Wallace SS. Characterization of Escherichia coli endonuclease VIII. J Biol Chem. 1997 Dec 19;272(51):32230-9. PMID:9405426
- ↑ Sidorenko VS, Rot MA, Filipenko ML, Nevinsky GA, Zharkov DO. Novel DNA glycosylases from Mycobacterium tuberculosis. Biochemistry (Mosc). 2008 Apr;73(4):442-50. PMID:18457574
- ↑ Kathe SD, Barrantes-Reynolds R, Jaruga P, Newton MR, Burrows CJ, Bandaru V, Dizdaroglu M, Bond JP, Wallace SS. Plant and fungal Fpg homologs are formamidopyrimidine DNA glycosylases but not 8-oxoguanine DNA glycosylases. DNA Repair (Amst). 2009 May 1;8(5):643-53. Epub 2009 Feb 12. PMID:19217358 doi:10.1016/j.dnarep.2008.12.013
- ↑ Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst). 2005 Jan 2;4(1):41-50. PMID:15533836 doi:10.1016/j.dnarep.2004.07.006
- ↑ Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst). 2003 May 13;2(5):581-91. PMID:12713815
- ↑ Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst). 2005 Jan 2;4(1):41-50. PMID:15533836 doi:10.1016/j.dnarep.2004.07.006
- ↑ Takao M, Oohata Y, Kitadokoro K, Kobayashi K, Iwai S, Yasui A, Yonei S, Zhang QM. Human Nei-like protein NEIL3 has AP lyase activity specific for single-stranded DNA and confers oxidative stress resistance in Escherichia coli mutant. Genes Cells. 2009 Feb;14(2):261-70. Epub 2008 Jan 15. PMID:19170771 doi:10.1111/j.1365-2443.2008.01271.x
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000 Aug 1;19(15):3857-69. PMID:10921868 doi:http://dx.doi.org/10.1093/emboj/19.15.3857
- ↑ Pereira de Jesus K, Serre L, Zelwer C, Castaing B. Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 2005 Oct 20;33(18):5936-44. Print 2005. PMID:16243784 doi:http://dx.doi.org/33/18/5936
- ↑ Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002 May 31;277(22):19811-6. Epub 2002 Mar 23. PMID:11912217 doi:http://dx.doi.org/10.1074/jbc.M202058200
- ↑ Pereira de Jesus K, Serre L, Zelwer C, Castaing B. Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 2005 Oct 20;33(18):5936-44. Print 2005. PMID:16243784 doi:http://dx.doi.org/33/18/5936
- ↑ Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J Biol Chem. 2003 Dec 19;278(51):51543-8. Epub 2003 Oct 1. PMID:14525999 doi:10.1074/jbc.M307768200
- ↑ Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000 Aug 1;19(15):3857-69. PMID:10921868 doi:http://dx.doi.org/10.1093/emboj/19.15.3857
- ↑ Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002 May 31;277(22):19811-6. Epub 2002 Mar 23. PMID:11912217 doi:http://dx.doi.org/10.1074/jbc.M202058200
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Imamura K, Wallace SS, Doublie S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J Biol Chem. 2009 Sep 18;284(38):26174-83. Epub 2009 Jul 22. PMID:19625256 doi:10.1074/jbc.M109.021907
- ↑ Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus. DNA Repair (Amst). 2007 Nov;6(11):1629-41. Epub 2007 Jul 12. PMID:17627905 doi:10.1016/j.dnarep.2007.05.011
- ↑ Coste F, Ober M, Carell T, Boiteux S, Zelwer C, Castaing B. Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase. J Biol Chem. 2004 Oct 15;279(42):44074-83. Epub 2004 Jul 10. PMID:15249553 doi:10.1074/jbc.M405928200
- ↑ Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci U S A. 2004 Jul 13;101(28):10284-9. Epub 2004 Jul 1. PMID:15232006 doi:10.1073/pnas.0402051101