Ann Taylor Sandbox 7
From Proteopedia
| (24 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | {{ | + | {{STRUCTURE_2ada| PDB=2ada | SCENE= }} |
| - | + | ||
| - | + | ===ADENOSINE DEAMINASE=== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ===Human p53 core domain with hot spot mutation R249S (I)=== | ||
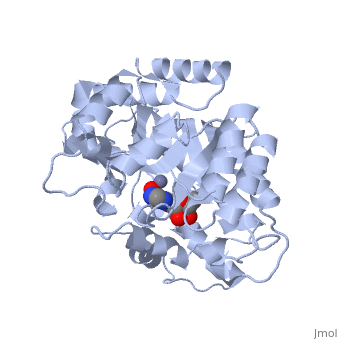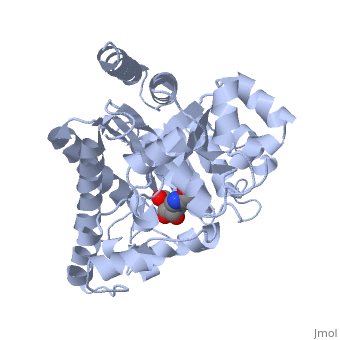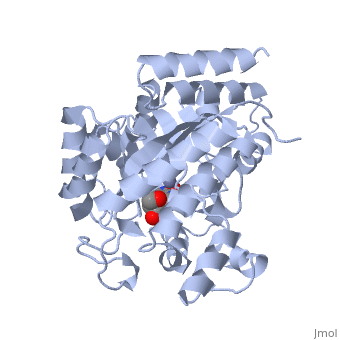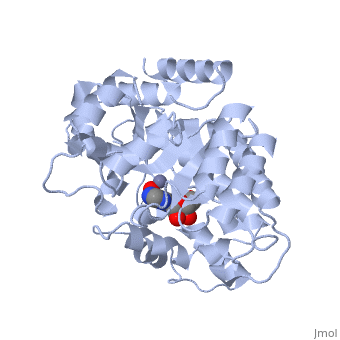
| + | Adenosine deaminase is involved in the degradation of purine nucleotides. It is especially active in lympocytes, and mutation of adenosine deaminase results in severe immunodeficiency. Adenosine deaminase contains an eight stranded parallel alpha/beta barrel with the active site in a deep pocket at the beta-barrel COOH-terminal end. <ref>PMID:1925539 </ref> The active site contains a <scene name='36/365340/Zinc_cofactor/1'>zinc cofactor</scene>, which coordinates to the 6-hydroxyl of the transition state analogue, 6-hydroxyl, 1,6-dihydropurine ribonucleoside. The <scene name='36/365340/Zinc_cofactor/1'>zinc</scene> is coordinated to three histidine residues and an aspartic acid residue. | ||
| - | The | + | The transition state analogue held in place mostly by polar interactions. The ribose group is close to the opening of the pocket, with the <scene name='36/365340/Purine_ring/1'>purine</scene> portion deeper in the pocket, close to the zinc. Nine hydrogen bonds stabilize the transition state-enzyme complex. |
| - | + | ADA is very stereoselective for the 6R isomer. This specificity is due to the location of the <scene name='36/365340/Zinc_cofactor/1'>catalytic zinc</scene>, <scene name='36/365340/Asp295/1'>Asp295</scene> and <scene name='36/365340/His_238/1'>His238</scene>. Interestingly, one face of the purine ring is exposed to polar groups and <scene name='36/365340/Zinc_cofactor/1'>zinc</scene>, while the other face is only exposed to nonpolar residues. The proposed catalytic mechanism has <scene name='36/365340/Asp295/1'>Asp295</scene> act as a general base, while the <scene name='36/365340/Zinc_cofactor/1'>zinc</scene> acts as an electrophile to activate the water molecule. <scene name='36/365340/Asp295/1'>Asp295</scene> and <scene name='36/365340/His_238/1'>His 238</scene> orients the water and stabilizes the charge of the attacking hydroxide. The protonated <scene name='36/365340/Glu217/1'>Glu217</scene> or the water hydrogen bonded to it could donate or share a proton with the N1 of the substrate. | |
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. | ||
| + | <references/> | ||
| - | ==Disease== | ||
| - | Known disease associated with this structure: Adrenal cortical carcinoma OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191170 191170]], Breast cancer OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191170 191170]], Colorectal cancer OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191170 191170]], Hepatocellular carcinoma OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191170 191170]], Histiocytoma OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191170 191170]], Li-Fraumeni syndrome OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191170 191170]], Multiple malignancy syndrome OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191170 191170]], Nasopharyngeal carcinoma OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191170 191170]], Osteosarcoma OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191170 191170]], Pancreatic cancer OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191170 191170]], Thyroid carcinoma OMIM:[[http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=191170 191170]] | ||
| - | ==About this Structure== | ||
| - | 3D06 is a 1 chain structure of sequence from [http://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3D06 OCA]. | ||
| - | + | <references/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
| |||||||||
| 2ada, resolution 2.40Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Activity: | Adenosine deaminase, with EC number 3.5.4.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
ADENOSINE DEAMINASE
Adenosine deaminase is involved in the degradation of purine nucleotides. It is especially active in lympocytes, and mutation of adenosine deaminase results in severe immunodeficiency. Adenosine deaminase contains an eight stranded parallel alpha/beta barrel with the active site in a deep pocket at the beta-barrel COOH-terminal end. [1] The active site contains a , which coordinates to the 6-hydroxyl of the transition state analogue, 6-hydroxyl, 1,6-dihydropurine ribonucleoside. The is coordinated to three histidine residues and an aspartic acid residue.
The transition state analogue held in place mostly by polar interactions. The ribose group is close to the opening of the pocket, with the portion deeper in the pocket, close to the zinc. Nine hydrogen bonds stabilize the transition state-enzyme complex.
ADA is very stereoselective for the 6R isomer. This specificity is due to the location of the , and . Interestingly, one face of the purine ring is exposed to polar groups and , while the other face is only exposed to nonpolar residues. The proposed catalytic mechanism has act as a general base, while the acts as an electrophile to activate the water molecule. and orients the water and stabilizes the charge of the attacking hydroxide. The protonated or the water hydrogen bonded to it could donate or share a proton with the N1 of the substrate.
- ↑ Wilson DK, Rudolph FB, Quiocho FA. Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science. 1991 May 31;252(5010):1278-84. PMID:1925539