We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
3i2t
From Proteopedia
(Difference between revisions)
| (10 intermediate revisions not shown.) | |||
| Line 1: | Line 1: | ||
| - | {{Seed}} | ||
| - | [[Image:3i2t.jpg|left|200px]] | ||
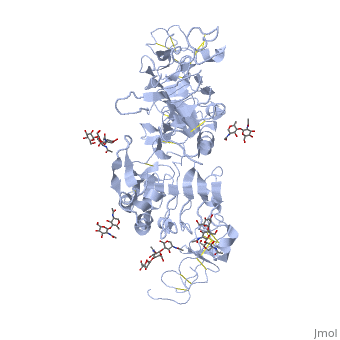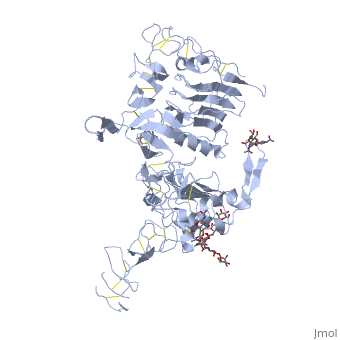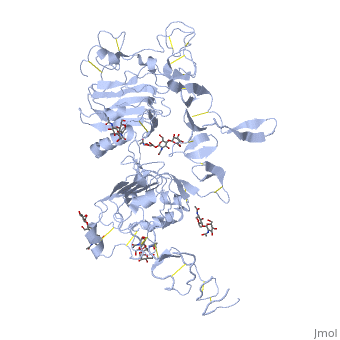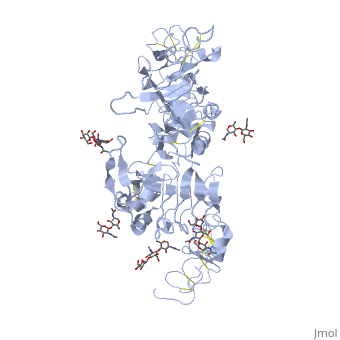
| - | < | + | ==Crystal structure of the unliganded Drosophila Epidermal Growth Factor Receptor ectodomain== |
| - | + | <StructureSection load='3i2t' size='340' side='right'caption='[[3i2t]], [[Resolution|resolution]] 2.70Å' scene=''> | |
| - | You may | + | == Structural highlights == |
| - | or the | + | <table><tr><td colspan='2'>[[3i2t]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Drosophila_melanogaster Drosophila melanogaster]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3I2T OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3I2T FirstGlance]. <br> |
| - | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.7Å</td></tr> | |
| - | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=BMA:BETA-D-MANNOSE'>BMA</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene>, <scene name='pdbligand=NDG:2-(ACETYLAMINO)-2-DEOXY-A-D-GLUCOPYRANOSE'>NDG</scene></td></tr> | |
| - | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3i2t FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3i2t OCA], [https://pdbe.org/3i2t PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3i2t RCSB], [https://www.ebi.ac.uk/pdbsum/3i2t PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3i2t ProSAT]</span></td></tr> | |
| + | </table> | ||
| + | == Function == | ||
| + | [https://www.uniprot.org/uniprot/EGFR_DROME EGFR_DROME] Binds to four ligands: Spitz, Gurken, Vein and Argos, which is an antagonist. Transduces the signal through the ras-raf-MAPK pathway. Involved in a myriad of developmental decisions. Critical for the proliferation of imaginal tissues, and for the determination of both the antero-posterior and dorso-ventral polarities of the oocyte. In the embryo, plays a role in the establishment of ventral cell fates, maintenance of amnioserosa and ventral neuroectodermal cells, germ band retraction, cell fate specification in the central nervous system and production of cuticle. Required for embryonic epithelial tissue repair.<ref>PMID:22140578</ref> | ||
| + | == Evolutionary Conservation == | ||
| + | [[Image:Consurf_key_small.gif|200px|right]] | ||
| + | Check<jmol> | ||
| + | <jmolCheckbox> | ||
| + | <scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/i2/3i2t_consurf.spt"</scriptWhenChecked> | ||
| + | <scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | ||
| + | <text>to colour the structure by Evolutionary Conservation</text> | ||
| + | </jmolCheckbox> | ||
| + | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=3i2t ConSurf]. | ||
| + | <div style="clear:both"></div> | ||
| + | <div style="background-color:#fffaf0;"> | ||
| + | == Publication Abstract from PubMed == | ||
| + | The orphan receptor tyrosine kinase ErbB2 (also known as HER2 or Neu) transforms cells when overexpressed, and it is an important therapeutic target in human cancer. Structural studies have suggested that the oncogenic (and ligand-independent) signalling properties of ErbB2 result from the absence of a key intramolecular 'tether' in the extracellular region that autoinhibits other human ErbB receptors, including the epidermal growth factor (EGF) receptor. Although ErbB2 is unique among the four human ErbB receptors, here we show that it is the closest structural relative of the single EGF receptor family member in Drosophila melanogaster (dEGFR). Genetic and biochemical data show that dEGFR is tightly regulated by growth factor ligands, yet a crystal structure shows that it, too, lacks the intramolecular tether seen in human EGFR, ErbB3 and ErbB4. Instead, a distinct set of autoinhibitory interdomain interactions hold unliganded dEGFR in an inactive state. All of these interactions are maintained (and even extended) in ErbB2, arguing against the suggestion that ErbB2 lacks autoinhibition. We therefore suggest that normal and pathogenic ErbB2 signalling may be regulated by ligands in the same way as dEGFR. Our findings have important implications for ErbB2 regulation in human cancer, and for developing therapeutic approaches that target novel aspects of this orphan receptor. | ||
| - | + | ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor.,Alvarado D, Klein DE, Lemmon MA Nature. 2009 Sep 10;461(7261):287-91. Epub 2009 Aug 30. PMID:19718021<ref>PMID:19718021</ref> | |
| + | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| + | </div> | ||
| + | <div class="pdbe-citations 3i2t" style="background-color:#fffaf0;"></div> | ||
| - | == | + | ==See Also== |
| - | + | *[[Epidermal Growth Factor Receptor|Epidermal Growth Factor Receptor]] | |
| + | *[[Epidermal growth factor receptor 3D structures|Epidermal growth factor receptor 3D structures]] | ||
| + | == References == | ||
| + | <references/> | ||
| + | __TOC__ | ||
| + | </StructureSection> | ||
[[Category: Drosophila melanogaster]] | [[Category: Drosophila melanogaster]] | ||
| - | [[Category: | + | [[Category: Large Structures]] |
| - | [[Category: | + | [[Category: Alvarado D]] |
| - | [[Category: | + | [[Category: Klein DE]] |
| - | [[Category: | + | [[Category: Lemmon MA]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Crystal structure of the unliganded Drosophila Epidermal Growth Factor Receptor ectodomain
| |||||||||||