User:Laura Fountain/Chloride Ion Channel
From Proteopedia
| Line 3: | Line 3: | ||
<applet load='1k0o' size='300' frame='true' align='right' caption='Soluble form of CLIC1' /> | <applet load='1k0o' size='300' frame='true' align='right' caption='Soluble form of CLIC1' /> | ||
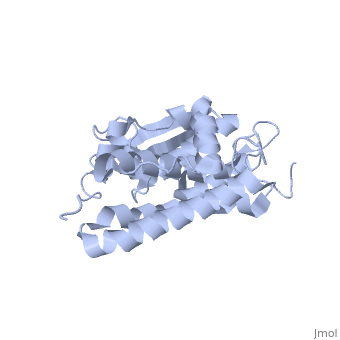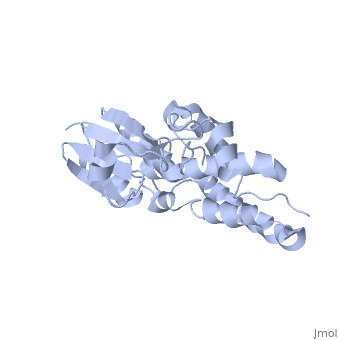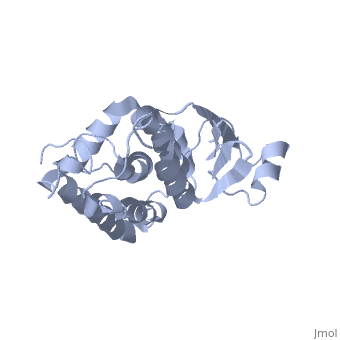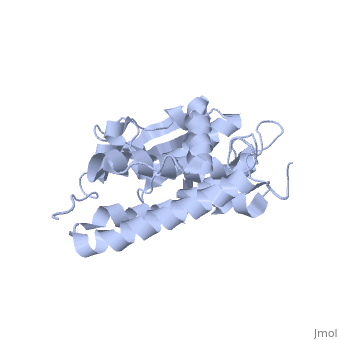
| - | CLIC1 (NCC27) is a member of the highly conserved class of chloride ion channels that exist in both soluble and integral membrane forms. | + | CLIC1 (NCC27) is a member of the highly conserved class of chloride ion channels that exist in both soluble and integral membrane forms. The CLIC family consists of seven distinct members: CLIC1, CLIC2, CLIC3, CLIC4, CLIC5, p64, and parchorin. The family is defined by a COOH-terminal core segment of ~230 amino acids that is highly conserved among all family members. CLIC1 has only a few amino acids upstream of this conserved core. CLIC1 is the most commonly studied member of the CLIC family because it is expressed to some extent in most tissues and cell types that have been studied and is particularly highly expressed in muscle.<ref>PMID:#11940526</ref> |
== About this Structure == | == About this Structure == | ||
| + | |||
| + | Purified CLIC1 can integrate into synthetic lipid bilayers forming a chloride channel with similar properties to those observed in vivo. The structure of the soluble form of CLIC1 has been determined at 1.4-A resolution, and is shown to the right. Integration of CLIC1 into the membrane is likely to require a major structural rearrangement, probably of the N-domain (<scene name='User:Laura_Fountain/Sandbox_1/N-domain/1'>residues 1-90</scene>), with the putative transmembrane helix arising from residues in the vicinity of the redox-active site.<ref>PMID:#11551966</ref> | ||
1K0O is a 2 chains structure of sequences from Homo sapiens. The protein is monomeric and structurally homologous to the glutathione S-transferase superfamily, and it has a redox-active site resembling glutaredoxin. The structure of the complex of CLIC1 with glutathione shows that glutathione occupies the redox-active site, which is adjacent to an open, elongated slot lined by basic residues. This structure indicates that CLIC1 is likely to be controlled by redox-dependent processes.<ref>PMID:#11551966</ref> | 1K0O is a 2 chains structure of sequences from Homo sapiens. The protein is monomeric and structurally homologous to the glutathione S-transferase superfamily, and it has a redox-active site resembling glutaredoxin. The structure of the complex of CLIC1 with glutathione shows that glutathione occupies the redox-active site, which is adjacent to an open, elongated slot lined by basic residues. This structure indicates that CLIC1 is likely to be controlled by redox-dependent processes.<ref>PMID:#11551966</ref> | ||
Revision as of 10:57, 30 September 2009
CLIC1: A Chloride Ion Channel
|
CLIC1 (NCC27) is a member of the highly conserved class of chloride ion channels that exist in both soluble and integral membrane forms. The CLIC family consists of seven distinct members: CLIC1, CLIC2, CLIC3, CLIC4, CLIC5, p64, and parchorin. The family is defined by a COOH-terminal core segment of ~230 amino acids that is highly conserved among all family members. CLIC1 has only a few amino acids upstream of this conserved core. CLIC1 is the most commonly studied member of the CLIC family because it is expressed to some extent in most tissues and cell types that have been studied and is particularly highly expressed in muscle.[1]
About this Structure
Purified CLIC1 can integrate into synthetic lipid bilayers forming a chloride channel with similar properties to those observed in vivo. The structure of the soluble form of CLIC1 has been determined at 1.4-A resolution, and is shown to the right. Integration of CLIC1 into the membrane is likely to require a major structural rearrangement, probably of the N-domain (), with the putative transmembrane helix arising from residues in the vicinity of the redox-active site.[2]
1K0O is a 2 chains structure of sequences from Homo sapiens. The protein is monomeric and structurally homologous to the glutathione S-transferase superfamily, and it has a redox-active site resembling glutaredoxin. The structure of the complex of CLIC1 with glutathione shows that glutathione occupies the redox-active site, which is adjacent to an open, elongated slot lined by basic residues. This structure indicates that CLIC1 is likely to be controlled by redox-dependent processes.[3]
References
[1]Harrop SJ, DeMaere MZ, Fairlie WD, Reztsova T, Valenzuela SM, Mazzanti M, Tonini R, Qiu MR, Jankova L, Warton K, Bauskin AR, Wu WM, Pankhurst S, Campbell TJ, Breit SN, Curmi PM. Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution. J Biol Chem. 2001 Nov 30;276(48):44993-5000. Epub 2001 Sep 10. PMID:11551966