User:Laura Fountain/Chloride Ion Channel
From Proteopedia
| Line 1: | Line 1: | ||
== CLIC1: A Chloride Ion Channel == | == CLIC1: A Chloride Ion Channel == | ||
| - | + | CLIC1 (NCC27) is a member of the highly conserved class of chloride ion channels that exist in both soluble and integral membrane forms. The CLIC family consists of seven distinct members: CLIC1, CLIC2, CLIC3, CLIC4, CLIC5, p64, and parchorin. The family is defined by a COOH-terminal core segment of ~230 amino acids that is highly conserved among all family members. CLIC1 has only a few amino acids upstream of this conserved core. CLIC1 is the most commonly studied member of the CLIC family because it is expressed to some extent in most tissues and cell types that have been studied and is particularly highly expressed in muscle.<ref name="Tulk">PMID:11940526</ref>CLIC1 has also been found in various intracellular membranes such as the mitochondrial, nuclear, and endoplasmic reticular membranes.<ref>PMID:#12202911</ref> | |
| - | + | ||
| - | CLIC1 (NCC27) is a member of the highly conserved class of chloride ion channels that exist in both soluble and integral membrane forms. The CLIC family consists of seven distinct members: CLIC1, CLIC2, CLIC3, CLIC4, CLIC5, p64, and parchorin. The family is defined by a COOH-terminal core segment of ~230 amino acids that is highly conserved among all family members. CLIC1 has only a few amino acids upstream of this conserved core. CLIC1 is the most commonly studied member of the CLIC family because it is expressed to some extent in most tissues and cell types that have been studied and is particularly highly expressed in muscle.<ref>PMID: | + | |
Because of their wide array of locations within the cell there is still a lot of research being done to discover their various functions within the cell. Some of the possibilities currently listed are: cell signaling, cell division, apoptosis, and, of course, ion flow regulation. | Because of their wide array of locations within the cell there is still a lot of research being done to discover their various functions within the cell. Some of the possibilities currently listed are: cell signaling, cell division, apoptosis, and, of course, ion flow regulation. | ||
| + | |||
== About this Structure == | == About this Structure == | ||
| + | |||
| + | <applet load='1k0o' size='300' frame='true' align='right' caption='Soluble form of CLIC1' /> | ||
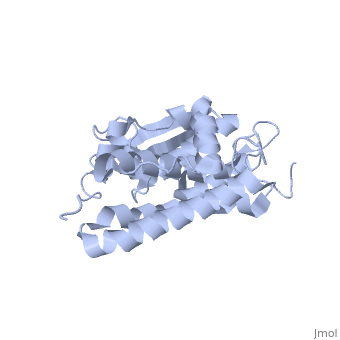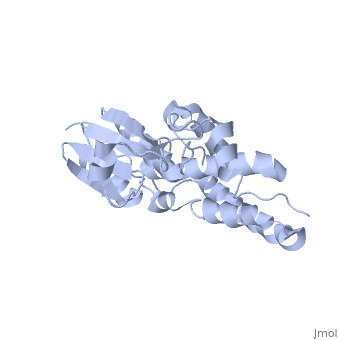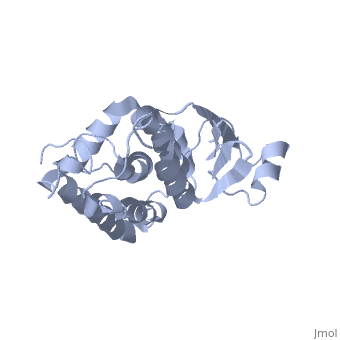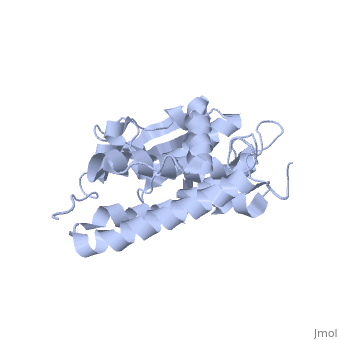
Purified CLIC1 can integrate into synthetic lipid bilayers forming a chloride channel with similar properties to those observed in vivo. The structure of the soluble form of CLIC1 has been determined at 1.4-A resolution, and is shown to the right. It's a homodimeric structure with one pore per subunit, creating a "double barreled" channel. At its binding site in the pore, chloride interacts with the ends of four helices that come from both sides of the membrane. A <scene name='User:Laura_Fountain/Sandbox_1/Glutamate_residue/1'>glutamate residue</scene> that protrudes into the pore is proposed to participate in gating.<ref>PMID:#12163078</ref> Integration of CLIC1 into the membrane is likely to require a major structural rearrangement, probably of the N-domain (<scene name='User:Laura_Fountain/Sandbox_1/N-domain/3'>residues 1-90</scene>), with the putative transmembrane helix arising from residues in the vicinity of the redox-active site.<ref>PMID:#11551966</ref> | Purified CLIC1 can integrate into synthetic lipid bilayers forming a chloride channel with similar properties to those observed in vivo. The structure of the soluble form of CLIC1 has been determined at 1.4-A resolution, and is shown to the right. It's a homodimeric structure with one pore per subunit, creating a "double barreled" channel. At its binding site in the pore, chloride interacts with the ends of four helices that come from both sides of the membrane. A <scene name='User:Laura_Fountain/Sandbox_1/Glutamate_residue/1'>glutamate residue</scene> that protrudes into the pore is proposed to participate in gating.<ref>PMID:#12163078</ref> Integration of CLIC1 into the membrane is likely to require a major structural rearrangement, probably of the N-domain (<scene name='User:Laura_Fountain/Sandbox_1/N-domain/3'>residues 1-90</scene>), with the putative transmembrane helix arising from residues in the vicinity of the redox-active site.<ref>PMID:#11551966</ref> | ||
| - | While this exact mechanism isn't known, it has been shown that functionality of the channel doesn't change whether it goes through 'normal' membrane integration via vesicles, or whether it's inserted into the intracellular space and allowed to integrate itself.<ref>PMID:#11940526</ref> Littler et. al. propose that upon oxidation CLIC1 undergoes a reversible transition from a monomeric to a non-covalent dimeric state due to the formation of an intramolecular disulfide bond (<scene name='User:Laura_Fountain/Sandbox_1/Cys_visualization/1'>Cys-24-Cys-59</scene>). They have determined the crystal structure of this oxidized state and show that a major structural transition has occurred, exposing a large hydrophobic surface, which forms the dimer interface. The oxidized CLIC1 dimer maintains its ability to form chloride ion channels in artificial bilayers and vesicles, whereas a reducing environment prevents the formation of ion channels by CLIC1. Their mutational studies show that both Cys-24 and Cys-59 are required for channel activity.<ref>PMID:#14613939</ref> | + | While this exact mechanism isn't known, it has been shown that functionality of the channel doesn't change whether it goes through 'normal' membrane integration via vesicles, or whether it's inserted into the intracellular space and allowed to integrate itself.<ref name="Tulk">PMID:#11940526</ref> Littler et. al. propose that upon oxidation CLIC1 undergoes a reversible transition from a monomeric to a non-covalent dimeric state due to the formation of an intramolecular disulfide bond (<scene name='User:Laura_Fountain/Sandbox_1/Cys_visualization/1'>Cys-24-Cys-59</scene>). They have determined the crystal structure of this oxidized state and show that a major structural transition has occurred, exposing a large hydrophobic surface, which forms the dimer interface. The oxidized CLIC1 dimer maintains its ability to form chloride ion channels in artificial bilayers and vesicles, whereas a reducing environment prevents the formation of ion channels by CLIC1. Their mutational studies show that both Cys-24 and Cys-59 are required for channel activity.<ref>PMID:#14613939</ref> |
1K0O is a 2 chains structure of sequences from Homo sapiens. The protein is monomeric and structurally homologous to the glutathione S-transferase superfamily, and it has a redox-active site resembling glutaredoxin. The structure of the complex of CLIC1 with glutathione shows that glutathione occupies the redox-active site, which is adjacent to an open, elongated slot lined by basic residues. This structure indicates that CLIC1 is likely to be controlled by redox-dependent processes.<ref>PMID:#11551966</ref> | 1K0O is a 2 chains structure of sequences from Homo sapiens. The protein is monomeric and structurally homologous to the glutathione S-transferase superfamily, and it has a redox-active site resembling glutaredoxin. The structure of the complex of CLIC1 with glutathione shows that glutathione occupies the redox-active site, which is adjacent to an open, elongated slot lined by basic residues. This structure indicates that CLIC1 is likely to be controlled by redox-dependent processes.<ref>PMID:#11551966</ref> | ||
Revision as of 03:39, 5 October 2009
CLIC1: A Chloride Ion Channel
CLIC1 (NCC27) is a member of the highly conserved class of chloride ion channels that exist in both soluble and integral membrane forms. The CLIC family consists of seven distinct members: CLIC1, CLIC2, CLIC3, CLIC4, CLIC5, p64, and parchorin. The family is defined by a COOH-terminal core segment of ~230 amino acids that is highly conserved among all family members. CLIC1 has only a few amino acids upstream of this conserved core. CLIC1 is the most commonly studied member of the CLIC family because it is expressed to some extent in most tissues and cell types that have been studied and is particularly highly expressed in muscle.[1]CLIC1 has also been found in various intracellular membranes such as the mitochondrial, nuclear, and endoplasmic reticular membranes.[2]
Because of their wide array of locations within the cell there is still a lot of research being done to discover their various functions within the cell. Some of the possibilities currently listed are: cell signaling, cell division, apoptosis, and, of course, ion flow regulation.
About this Structure
|
Purified CLIC1 can integrate into synthetic lipid bilayers forming a chloride channel with similar properties to those observed in vivo. The structure of the soluble form of CLIC1 has been determined at 1.4-A resolution, and is shown to the right. It's a homodimeric structure with one pore per subunit, creating a "double barreled" channel. At its binding site in the pore, chloride interacts with the ends of four helices that come from both sides of the membrane. A that protrudes into the pore is proposed to participate in gating.[3] Integration of CLIC1 into the membrane is likely to require a major structural rearrangement, probably of the N-domain (), with the putative transmembrane helix arising from residues in the vicinity of the redox-active site.[4]
While this exact mechanism isn't known, it has been shown that functionality of the channel doesn't change whether it goes through 'normal' membrane integration via vesicles, or whether it's inserted into the intracellular space and allowed to integrate itself.[1] Littler et. al. propose that upon oxidation CLIC1 undergoes a reversible transition from a monomeric to a non-covalent dimeric state due to the formation of an intramolecular disulfide bond (). They have determined the crystal structure of this oxidized state and show that a major structural transition has occurred, exposing a large hydrophobic surface, which forms the dimer interface. The oxidized CLIC1 dimer maintains its ability to form chloride ion channels in artificial bilayers and vesicles, whereas a reducing environment prevents the formation of ion channels by CLIC1. Their mutational studies show that both Cys-24 and Cys-59 are required for channel activity.[5]
1K0O is a 2 chains structure of sequences from Homo sapiens. The protein is monomeric and structurally homologous to the glutathione S-transferase superfamily, and it has a redox-active site resembling glutaredoxin. The structure of the complex of CLIC1 with glutathione shows that glutathione occupies the redox-active site, which is adjacent to an open, elongated slot lined by basic residues. This structure indicates that CLIC1 is likely to be controlled by redox-dependent processes.[6]
References
- ↑ 1.0 1.1 Tulk BM, Kapadia S, Edwards JC. CLIC1 inserts from the aqueous phase into phospholipid membranes, where it functions as an anion channel. Am J Physiol Cell Physiol. 2002 May;282(5):C1103-12. PMID:11940526 doi:10.1152/ajpcell.00402.2001
- ↑ PMID:#12202911
- ↑ PMID:#12163078
- ↑ PMID:#11551966
- ↑ PMID:#14613939
- ↑ PMID:#11551966