User:Eleanor Crabb/Sandbox 1
From Proteopedia
| Line 1: | Line 1: | ||
| - | <applet load='1z3a' size='300' frame='true' align='right' caption=' | + | |
| - | The interactive image on the right shows the '''asymmetric unit''' of | + | == Visualising Protein Structure == |
| + | <applet load='1z3a' size='300' frame='true' align='right' caption='1z3a' />This page is designed to be an introduction to protein structure and the 3D visualisation package, '''Jmol''', used here in Proteopedia and also on the RCSB Protein Data Bank website to visualise proteins. | ||
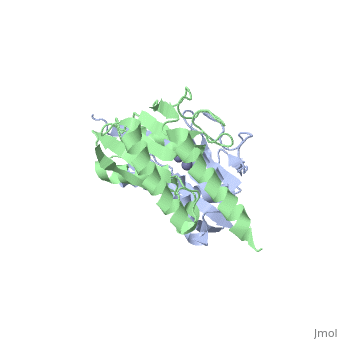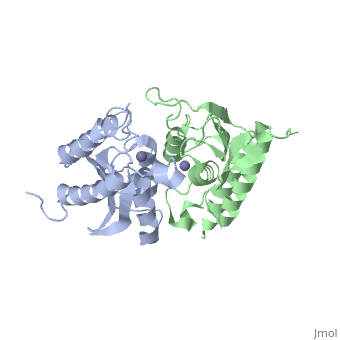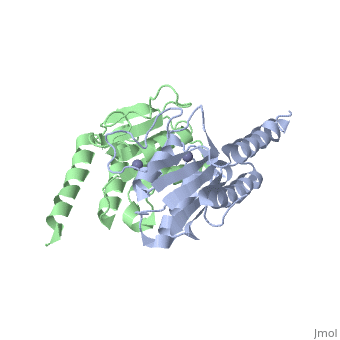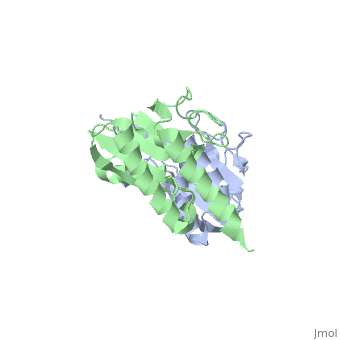
| + | The interactive image on the right shows the '''asymmetric unit''' of a zinc-containing enzyme with the pdb code '''1z3a'''. (The asymmetric unit is the smallest part of the crystal structure that can be used to generate the unit cell of a crystal and hence the entire crystal structure of a protein - see the asymmetric tutorial at the RCSB Protein Data Bank [http://www.rcsb.org/pdb/static.do?p=education_discussion/Looking-at-Structures/biounit_tutorial.html#Anchor-Asym] for more information.) | ||
| - | You can rotate the molecule using your mouse and zoom in and out by holding down the shift key while you move the mouse. You can also spin the molecule use the Toggle spin button below the molecule. | + | You can rotate the molecule using your mouse and zoom in and out by holding down the shift key while you move the mouse. You can also spin the molecule use the Toggle spin button below the molecule. (The popup button will open a resizable Jmol representation of the molecule in another window.) |
| - | You will note that there are two chains in this protein, shown in green and blue. For simplicity let us concentrate on just a <scene name='User:Eleanor_Crabb/Sandbox_1/1z3a_single_chain/1'>single chain</scene>, chain A. To see just chain A, click on the link in green. If you hover the mouse over a particular part of the structure, for example, the central zinc atom, you will see a yellow box with a label giving the atom identity. In this example, | + | You will note that there are two chains in this protein, shown in green and blue. For simplicity let us concentrate on just a <scene name='User:Eleanor_Crabb/Sandbox_1/1z3a_single_chain/1'>single chain</scene>, chain A. To see just chain A, click on the link in green. If you hover the mouse over a particular part of the structure, for example, the central zinc atom, you will see a yellow box with a label giving the atom identity. In this example, you will see the label [[Image:elementbox.JPG]]. Hovering the cursor over the backbone of the protein will show the constituent residues in the chain. |
This is a cartoon representation of the protein, showing a schematic representation of the back bone of the protein. You may be more familiar with the following molecular representations (note in all cases that the H atoms in the molecule are not shown): | This is a cartoon representation of the protein, showing a schematic representation of the back bone of the protein. You may be more familiar with the following molecular representations (note in all cases that the H atoms in the molecule are not shown): | ||
| - | *<scene name='User:Eleanor_Crabb/Sandbox_1/1z3a_a_sfllcolour/2'>space- filled</scene> - this gives an indication of the size and shape of the protein | + | *<scene name='User:Eleanor_Crabb/Sandbox_1/1z3a_a_sfllcolour/2'>space-filled</scene> - this gives an indication of the size and shape of the protein |
| - | *<scene name='User:Eleanor_Crabb/Sandbox_1/1z3a_a_ballcolour/1'>ball and stick</scene> - this view allows you to see | + | *<scene name='User:Eleanor_Crabb/Sandbox_1/1z3a_a_ballcolour/1'>ball and stick</scene> - this view allows you to see the atoms in the protein. |
Now, let us simplify things further by looking at just a | Now, let us simplify things further by looking at just a | ||
| Line 15: | Line 17: | ||
<scene name='User:Eleanor_Crabb/Sandbox_1/1z3a_single_helix/1'>cartoon</scene> alone. | <scene name='User:Eleanor_Crabb/Sandbox_1/1z3a_single_helix/1'>cartoon</scene> alone. | ||
| - | + | [[Eleanor_Crabb/The_four_tiers_of_protein_structure|Next page - The four tiers of protein structure]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Visualising Protein Structure
|
The interactive image on the right shows the asymmetric unit of a zinc-containing enzyme with the pdb code 1z3a. (The asymmetric unit is the smallest part of the crystal structure that can be used to generate the unit cell of a crystal and hence the entire crystal structure of a protein - see the asymmetric tutorial at the RCSB Protein Data Bank [1] for more information.)
You can rotate the molecule using your mouse and zoom in and out by holding down the shift key while you move the mouse. You can also spin the molecule use the Toggle spin button below the molecule. (The popup button will open a resizable Jmol representation of the molecule in another window.)
You will note that there are two chains in this protein, shown in green and blue. For simplicity let us concentrate on just a , chain A. To see just chain A, click on the link in green. If you hover the mouse over a particular part of the structure, for example, the central zinc atom, you will see a yellow box with a label giving the atom identity. In this example, you will see the label . Hovering the cursor over the backbone of the protein will show the constituent residues in the chain.
This is a cartoon representation of the protein, showing a schematic representation of the back bone of the protein. You may be more familiar with the following molecular representations (note in all cases that the H atoms in the molecule are not shown):
- - this gives an indication of the size and shape of the protein
- - this view allows you to see the atoms in the protein.
Now, let us simplify things further by looking at just a of the chain. If you rotate the model you can clearly see the backbone linking the different residues. We now draw a line through the backbone of the chain illustrated as or the alone.