User:Stéphanie Kraemer/Sandbox 106
From Proteopedia
(→'''Structure and function''') |
|||
| Line 17: | Line 17: | ||
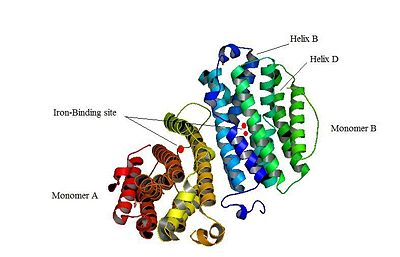
<scene name='User:Stéphanie_Kraemer/Sandbox_106/Iron-binding_site/2'>An iron-binding site</scene>is highlighted. But concerning this site, the two monomers are not the same. Actually, the B monomer has two iron-binding site (called Fe2 and Fe1) whereas the A monomer has only one which is Fe2. This can be explained by structural changes in the helix that compose the two monomers. The 37 to 42 N-terminal residues (called the swivel loop) from one monomer can rotate between two conformations and can influence the position of the helix B or D on the opposite monomer. | <scene name='User:Stéphanie_Kraemer/Sandbox_106/Iron-binding_site/2'>An iron-binding site</scene>is highlighted. But concerning this site, the two monomers are not the same. Actually, the B monomer has two iron-binding site (called Fe2 and Fe1) whereas the A monomer has only one which is Fe2. This can be explained by structural changes in the helix that compose the two monomers. The 37 to 42 N-terminal residues (called the swivel loop) from one monomer can rotate between two conformations and can influence the position of the helix B or D on the opposite monomer. | ||
| - | [[Image: | + | [[Image:Thetwomonomers.jpg|400px|thumb]] |
Revision as of 12:50, 16 November 2009
p53R2
| |||||||
| 3hf1, resolution 2.60Å () | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||
| Gene: | RRM2B, P53R2 (Homo sapiens) | ||||||
| Activity: | Ribonucleoside-diphosphate reductase, with EC number 1.17.4.1 | ||||||
| Related: | 1xsm, 2uw2, 1w69, 1w68 | ||||||
| |||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
P53R2 is an oxydoreductase composed of 351 residues. It is a small subunit of the ribonucleotide reductase (RNR). RNR catalyses the reduction of the four nucleotides to desoxyribonucleotides. It exists three classes of RNR. Class I RNR is a tetramer composed of the two types of subunits with stoichiometry α2β2 and three subunits have been identified in mammals :
- Large (α) subunit M1 that contains the enzyme active site
- Small (β) subunit M2 that contains a dinuclear iron site, it’s the regulatory subunit
- P53R2, the last identified (in 2000), which is transactivated by p53 in response to DNA damage in cells during the G0-G1 cell cycle phase
M2 and p53R2 interact with M1 through the C-terminal binding domain. These two subunits share more than 80% sequence identity. But the few differences between the two are not unimportant, as it’s explained below. The first X-ray crystal structure of p53R2 has a resolution of 2,6 Å and permits to describe its structure and also to show the structural differences with the M2 subunit.
Structure and function
The X-ray crystal structure permits to see that p53R2 is made of two monomers A and B themselves made of loops and helix. Two of them play an important role. is highlighted. But concerning this site, the two monomers are not the same. Actually, the B monomer has two iron-binding site (called Fe2 and Fe1) whereas the A monomer has only one which is Fe2. This can be explained by structural changes in the helix that compose the two monomers. The 37 to 42 N-terminal residues (called the swivel loop) from one monomer can rotate between two conformations and can influence the position of the helix B or D on the opposite monomer.