Glycogen Phosphorylase
From Proteopedia
| Line 1: | Line 1: | ||
<table style="background-color:#ffffc0" cellpadding="8" width="95%" border="0"><tr><td>Please do NOT make changes to this Sandbox until after April 23, 2010. Sandboxes 151-200 are reserved until then for use by the Chemistry 307 class at UNBC taught by Prof. [[User:Andrea Gorrell|Andrea Gorrell]].</td></tr> | <table style="background-color:#ffffc0" cellpadding="8" width="95%" border="0"><tr><td>Please do NOT make changes to this Sandbox until after April 23, 2010. Sandboxes 151-200 are reserved until then for use by the Chemistry 307 class at UNBC taught by Prof. [[User:Andrea Gorrell|Andrea Gorrell]].</td></tr> | ||
| - | + | ||
'''Glycogen Phosphorylase<br> | '''Glycogen Phosphorylase<br> | ||
{{STRUCTURE_1z6q| PDB=1z6q | SCENE= }} | {{STRUCTURE_1z6q| PDB=1z6q | SCENE= }} | ||
| - | =Introduction | + | =Introduction= |
| - | Glycogen phosphorylase catalyzes the hydrolysis of glycogen to generate glucose-1-phosphate and is considered the rate limiting step in the degradation of glycogen. The glucose-1-phophate is then further degraded via the pathway of glycolysis. There is both muscle and liver glycogen phosphorylase; muscle glycogen phosphorylase is present to degrade glycogen to forms of energy by means of glycolysis during muscle contractions and liver glycogen is present to regulate the blood glucose levels within the blood. | + | Glycogen phosphorylase catalyzes the hydrolysis of glycogen to generate glucose-1-phosphate and shortened glycogen molecule and is considered the rate limiting step in the degradation of glycogen<ref name="gp">PMID: 15214781 </ref>. It is a part of the glucosyltransferase family and acts on the α-1,4-glycosidic linkages. The phosphorylase comes to a standstill 4 residues from an α-1,6-branchpoint, where debranching enzyme takes over <ref name =“gp3”> PMID: 11949930</ref>. The glucose-1-phophate is then further degraded via the pathway of glycolysis. There is both muscle and liver glycogen phosphorylase; muscle glycogen phosphorylase is present to degrade glycogen to forms of energy by means of glycolysis during muscle contractions and liver glycogen is present to regulate the blood glucose levels within the blood <ref name =“gp3”/>. |
=Structure and Function= | =Structure and Function= | ||
| - | Glycogen phosphorylase is a dimer consisting of two identical subunits and has an essential cofactor, pryridoxal phosphate (PLP). Glycogen phosphorylase can be found in two different states, glycogen phosphorylase a (GP''a'') and glycogen phosphorylase b (GP''b'') | + | Glycogen phosphorylase is a dimer consisting of two identical subunits and has an essential cofactor, pryridoxal phosphate (PLP). Glycogen phosphorylase can be found in two different states, glycogen phosphorylase a (GP''a'') and glycogen phosphorylase b (GP''b'')<ref name="gp"/>The difference in the structures is due to phosphorylation of the <scene name='Sandbox_153/Ser14/1'>Ser-14</scene> residue which results in the active form (GP''a''). Protein phosphatases dephosphorylate the GP''a'' to the inactive form, also known as GP''b''. Both forms of glycogen phosphorylase can also be found in T and R states where T is the inactive state because it appears to have a low affinity for substrate and R is the active state where it appears to have a greater affinity for substrate<ref name="gp2">PMID: 1900534</ref>. The secondary structures of T and R states of GP''a'' and ''b'' are similar with an <scene name='Sandbox_153/Nterminaldomain/2'>N-terminal domain </scene> and a <scene name='Sandbox_153/Cterminaldomain/1'>C-terminal domain </scene> <ref name"">DOI: 10.1021/ar00083a004 </ref>. Each domain also contains subdomains which undergo conformational changes on the interconversion of T and R states <ref name="gp2"/>. The R states of GP''a'' and GP''b'' are almost identical; the difference lays in the modification of the Ser-14 residue where GP''a'' has a covalently linked phosphate group whereas GP''b'' has a non-covalently linked sulfide group . GP''a'' is activated by phosphorylation of the serine residue whereas GP''b'' can be activated by the binding of AMP to the allosteric site that is present within the molecule. GP''a'' does not require the binding of AMP but attachment enhances the activity of the enzyme upwards to 25%. |
<scene name='Sandbox_153/Activesites/3'>active sites of glycogen phosphorylase </scene> | <scene name='Sandbox_153/Activesites/3'>active sites of glycogen phosphorylase </scene> | ||
<scene name='Sandbox_153/Catalyticsite/1'> catalytic site </scene> | <scene name='Sandbox_153/Catalyticsite/1'> catalytic site </scene> | ||
(ADD IN REFERENCES) | (ADD IN REFERENCES) | ||
=Mechanism= | =Mechanism= | ||
| - | In muscle, glycogen phosphorylase is activated by hormones and neural signals such as epinephrine, stimulate phosphorylase kinase which phosphorylates the Ser-14 residue of the protein. A second messenger of cyclic AMP (cAMP) increases in concentration due to epinephrine or glucagon, and this increase results in an enzyme | + | In muscle, glycogen phosphorylase is activated by hormones and neural signals such as epinephrine, that stimulate phosphorylase kinase which phosphorylates the Ser-14 residue of the protein. A second messenger of cyclic AMP (cAMP) increases in concentration due to epinephrine or glucagon, and this increase results in an enzyme cascade. [[Image:Enzyme cascade.jpg|thumb|alt=enzyme cascade|Figure 1: Diagram illustrating the enzyme cascade that occurs when hormones(and neural signals)catalyze the activation of phosphorylase through kinase activity and thus inactivating glycogen synthetase. Phosphatases inactivate phosphorylase and is regulated by phosphorylase itself as well as inhibitor proteins. High glucose concentratios also lead to the inactivation of phosphorylase (inactivation represented by open arrows). Diagram adapted from (Ref).]]Protein kinase A is also activated by this increase in cAMP which then phosphorylates phosphorylase kinase, which in turn activates the glycogen phosphorylase enzyme by phosphorylating the Ser-14 residue. In the liver, glucagon is the primary signal which catalyzes this enzyme cascade. (ADD IN REFERENCES) |
=References= | =References= | ||
<references/> | <references/> | ||
Revision as of 20:17, 30 March 2010
| Please do NOT make changes to this Sandbox until after April 23, 2010. Sandboxes 151-200 are reserved until then for use by the Chemistry 307 class at UNBC taught by Prof. Andrea Gorrell. |
Contents |
Introduction
Glycogen phosphorylase catalyzes the hydrolysis of glycogen to generate glucose-1-phosphate and shortened glycogen molecule and is considered the rate limiting step in the degradation of glycogen[1]. It is a part of the glucosyltransferase family and acts on the α-1,4-glycosidic linkages. The phosphorylase comes to a standstill 4 residues from an α-1,6-branchpoint, where debranching enzyme takes over [2]. The glucose-1-phophate is then further degraded via the pathway of glycolysis. There is both muscle and liver glycogen phosphorylase; muscle glycogen phosphorylase is present to degrade glycogen to forms of energy by means of glycolysis during muscle contractions and liver glycogen is present to regulate the blood glucose levels within the blood [2].
Structure and Function
Glycogen phosphorylase is a dimer consisting of two identical subunits and has an essential cofactor, pryridoxal phosphate (PLP). Glycogen phosphorylase can be found in two different states, glycogen phosphorylase a (GPa) and glycogen phosphorylase b (GPb)[1]The difference in the structures is due to phosphorylation of the residue which results in the active form (GPa). Protein phosphatases dephosphorylate the GPa to the inactive form, also known as GPb. Both forms of glycogen phosphorylase can also be found in T and R states where T is the inactive state because it appears to have a low affinity for substrate and R is the active state where it appears to have a greater affinity for substrate[3]. The secondary structures of T and R states of GPa and b are similar with an and a [4]. Each domain also contains subdomains which undergo conformational changes on the interconversion of T and R states [3]. The R states of GPa and GPb are almost identical; the difference lays in the modification of the Ser-14 residue where GPa has a covalently linked phosphate group whereas GPb has a non-covalently linked sulfide group . GPa is activated by phosphorylation of the serine residue whereas GPb can be activated by the binding of AMP to the allosteric site that is present within the molecule. GPa does not require the binding of AMP but attachment enhances the activity of the enzyme upwards to 25%. (ADD IN REFERENCES)
Mechanism
In muscle, glycogen phosphorylase is activated by hormones and neural signals such as epinephrine, that stimulate phosphorylase kinase which phosphorylates the Ser-14 residue of the protein. A second messenger of cyclic AMP (cAMP) increases in concentration due to epinephrine or glucagon, and this increase results in an enzyme cascade.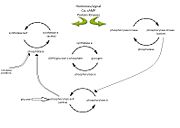
References
- ↑ 1.0 1.1 Kristiansen M, Andersen B, Iversen LF, Westergaard N. Identification, synthesis, and characterization of new glycogen phosphorylase inhibitors binding to the allosteric AMP site. J Med Chem. 2004 Jul 1;47(14):3537-45. PMID:15214781 doi:10.1021/jm031121n
- ↑ 2.0 2.1 Roach PJ. Glycogen and its metabolism. Curr Mol Med. 2002 Mar;2(2):101-20. PMID:11949930
- ↑ 3.0 3.1 Barford D, Hu SH, Johnson LN. Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP. J Mol Biol. 1991 Mar 5;218(1):233-60. PMID:1900534
- ↑ doi: https://dx.doi.org/10.1021/ar00083a004
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Amy Chahal, Ann Taylor, Alexander Berchansky, Joel L. Sussman, Riley Hicks, Andrea Gorrell, David Canner
