Glycerol-3-Phosphate Dehydrogenase
From Proteopedia
| Line 2: | Line 2: | ||
'''Glycerol 3-Phosphate Dehydrogenase''' | '''Glycerol 3-Phosphate Dehydrogenase''' | ||
| - | Glycerol 3-phosphate dehydrogenase (GlpD) is a membrane bound enzyme in prokaryotes and in eukaryotes. Glycerol 3-Phosphate Dehydrogenase (GlpD) is an oxidoreductase enzyme which catalyzes the reduction in [http://en.wikipedia.org/wiki/File:Dihydroxyacetone_phosphate_to_glycerol_3-phosphate_en.svg reaction] of Dihydroxyacetone Phosphate to Glycerol 3-Phosphate. GlpD is involved in many cellular functions such as phospholipids biosynthesis, respiration and metabolism. The GlpD is a dimer consisting of two subunits which contain the | + | Glycerol 3-phosphate dehydrogenase (GlpD) is a membrane bound enzyme in prokaryotes and in eukaryotes. Glycerol 3-Phosphate Dehydrogenase (GlpD) is an oxidoreductase enzyme which catalyzes the reduction in [http://en.wikipedia.org/wiki/File:Dihydroxyacetone_phosphate_to_glycerol_3-phosphate_en.svg reaction] of Dihydroxyacetone Phosphate to Glycerol 3-Phosphate. GlpD is involved in many cellular functions such as phospholipids biosynthesis, respiration and metabolism. The GlpD is a dimer consisting of two subunits which contain the catabolite activator protein (CAP)-Domain,the flavin adenine dinucleotide(FAD)-Domain and a ubiquinone analogue, MD. |
===Structure=== | ===Structure=== | ||
| Line 10: | Line 10: | ||
<scene name='Sandbox_189/Cap_domain/1'>Cap-Binding Domain</scene> | <scene name='Sandbox_189/Cap_domain/1'>Cap-Binding Domain</scene> | ||
| - | The C-terminal consists of negatively charged residues that are opposite in orientation to the positively charged residues of the FAD-Domain in the phospholipid membrane. | + | The C-terminal CAP-Domain consists of negatively charged residues that are opposite in orientation to the positively charged residues of the FAD-Domain in the phospholipid membrane. The CAP-domain is responsible in gene transcription and helical turns. |
<scene name='Sandbox_189/Fad/2'>FAD Active Site</scene> | <scene name='Sandbox_189/Fad/2'>FAD Active Site</scene> | ||
| - | The N-terminal FAD-Domain exists in each monomer subunit of GlpD and is embedded into the phospholipid membrane bilayer. Substrate binding occurs at this domain which causes a conformational change to the structure of the GlpD enzyme. The | + | The N-terminal FAD-Domain exists in each monomer subunit of GlpD and is embedded into the phospholipid membrane bilayer.Substrate binding occurs at this domain which causes a conformational change to the structure of the GlpD enzyme. The FAD-domain plays a major role in metabolism and energy synthesis. |
===Function=== | ===Function=== | ||
| - | GlpD functions in the intracellular membrane of E. coli and in the inner-mitochondrial membrane of eukaryotes. In E. Coli, GlpD catalyzes and reduces the reaction of dihydroxyacetone phosphate to glycerol 3-phosphate in the [http://www.pnas.org/content/105/9/3280/F1.large.jpg glycerol metabolism pathway]. The binding of the substrate analogues and | + | GlpD functions in the intracellular membrane of E. coli and in the inner-mitochondrial membrane of eukaryotes. In E. Coli, GlpD catalyzes and reduces the reaction of dihydroxyacetone phosphate to glycerol 3-phosphate in the [http://www.pnas.org/content/105/9/3280/F1.large.jpg glycerol metabolism pathway]. The binding of the substrate analogues (glyceraldehydes 3-phosphate, glyceric acid 2-phosphate and phosphoenolpyruvate, dihydroxyacetone phosphate)or UQ substrate analogues (2-n-heptyl-4-hydroxyquinoline N-oxide and menadione). The conformational change of the structure and resiudes of GlpD catalyzes many different metabolic reactions. |
| + | |||
| + | ====Glycerol Metabolic Pathway==== | ||
| - | Upon the oxidation of glycerol 3-phosphate, flavin adenine dinucleotide (FAD) reduces to FADH2, passing on electrons to Ubiquinone(UQ). UQ then reduces to UQH2 which allows for the transport of electrons into the respiratory pathway. | ||
| - | |||
| - | Glycerol 3-phosphate dehydrogenase (GlpD) is a membrane bound enzyme in prokaryotes and in eukaryotes. GlpD is involved in many cellular functions, some of which are phospholipids biosynthesis, respiration and metabolism. The authors believed that GlpD undergoes a conformational change upon complexing with analogue substrates, which are thought to catalyze glycerol 3-phosphate (G3P) dehydrogenation in two possible ways. The authors further researched and discovered more GlpD structures that are bound to Ubiquonone (Ub) analogues in order to link catalytic dehydrogenation to respiration and to gain insight on the mechanism involved in the transport of electrons into the respiratory pathway. It is also thought by the authors that the prokaryotic enzyme structural results can be applied to eukaryotic GlpD enzyme structural results, due to the conservation of greater than 45% of consensus protein sequences in almost all organisms. | ||
| - | ====Metabolism==== | ||
===Diseases=== | ===Diseases=== | ||
===References=== | ===References=== | ||
Revision as of 06:02, 1 April 2010
Template:STRUCTURE 2r4e Glycerol 3-Phosphate Dehydrogenase
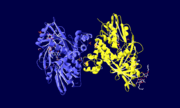
Glycerol 3-phosphate dehydrogenase (GlpD) is a membrane bound enzyme in prokaryotes and in eukaryotes. Glycerol 3-Phosphate Dehydrogenase (GlpD) is an oxidoreductase enzyme which catalyzes the reduction in reaction of Dihydroxyacetone Phosphate to Glycerol 3-Phosphate. GlpD is involved in many cellular functions such as phospholipids biosynthesis, respiration and metabolism. The GlpD is a dimer consisting of two subunits which contain the catabolite activator protein (CAP)-Domain,the flavin adenine dinucleotide(FAD)-Domain and a ubiquinone analogue, MD.
Contents |
Structure
GlpD is a dimer that consists of two subunits; α and β. The GlpD structure also contains seven ligands; 1,3-Dihydroxyacetonephosphate (13P), β-Octylglucoside (βOG), 1,2-Ethanediol (EDO), Flavin-Adenine Dinucleotide (FAD), Imidazole (IMD), PO4 (Phosphate Ion) and N-(Tris(Hydroxymethyl)methyl)-3-Aminopropanesulfonic Acid (T3A). The active sites on GlpD are the Cap-Domain, FAD- Domain and a ubiquinone substrate analogue, menadione (MD).
The C-terminal CAP-Domain consists of negatively charged residues that are opposite in orientation to the positively charged residues of the FAD-Domain in the phospholipid membrane. The CAP-domain is responsible in gene transcription and helical turns.
The N-terminal FAD-Domain exists in each monomer subunit of GlpD and is embedded into the phospholipid membrane bilayer.Substrate binding occurs at this domain which causes a conformational change to the structure of the GlpD enzyme. The FAD-domain plays a major role in metabolism and energy synthesis.
Function
GlpD functions in the intracellular membrane of E. coli and in the inner-mitochondrial membrane of eukaryotes. In E. Coli, GlpD catalyzes and reduces the reaction of dihydroxyacetone phosphate to glycerol 3-phosphate in the glycerol metabolism pathway. The binding of the substrate analogues (glyceraldehydes 3-phosphate, glyceric acid 2-phosphate and phosphoenolpyruvate, dihydroxyacetone phosphate)or UQ substrate analogues (2-n-heptyl-4-hydroxyquinoline N-oxide and menadione). The conformational change of the structure and resiudes of GlpD catalyzes many different metabolic reactions.
Glycerol Metabolic Pathway
Diseases
References
| Please do NOT make changes to this Sandbox until after April 23, 2010. Sandboxes 151-200 are reserved until then for use by the Chemistry 307 class at UNBC taught by Prof. Andrea Gorrell. |
Proteopedia Page Contributors and Editors (what is this?)
Indu Toora, Michal Harel, Alexander Berchansky, David Canner, Andrea Gorrell, Andrew Rebeyka, Jaime Prilusky