User:Wayne Decatur/Sandbox Glutamate receptor
From Proteopedia
m |
m |
||
| Line 3: | Line 3: | ||
==Structure of the Glutamate Receptor (GluA2)== | ==Structure of the Glutamate Receptor (GluA2)== | ||
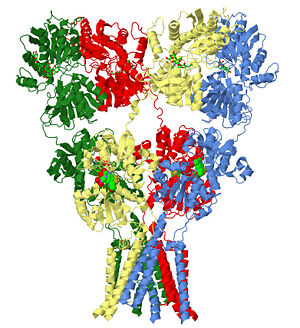
| - | The homomeric rat GluA2 receptor <scene name='User:Wayne_Decatur/Sandbox_Glutamate_receptor/Default3kg2/1'>has four subunits</scene> | + | The homomeric rat GluA2 receptor <scene name='User:Wayne_Decatur/Sandbox_Glutamate_receptor/Default3kg2/1'>has four subunits</scene> arranged in a 'Y'-shape with the <scene name='User:Wayne_Decatur/Sandbox_Glutamate_receptor/Meas3kg2/1'>'top' being about three times the width of the 'bottom'</scene><ref>PMID: 19946266</ref>. {{Link Toggle FancyCartoonHighQualityView}}. |
{{Structure | {{Structure | ||
|PDB= 3kg2 |SIZE=400|SCENE=User:Wayne_Decatur/Sandbox_Glutamate_receptor/Default3kg2/1|CAPTION= glutamate receptor ([[3kg2]]), resolution 3.6Å (<scene name='User:Wayne_Decatur/Sandbox_Glutamate_receptor/Default3kg2/1'>initial scene</scene>) | |PDB= 3kg2 |SIZE=400|SCENE=User:Wayne_Decatur/Sandbox_Glutamate_receptor/Default3kg2/1|CAPTION= glutamate receptor ([[3kg2]]), resolution 3.6Å (<scene name='User:Wayne_Decatur/Sandbox_Glutamate_receptor/Default3kg2/1'>initial scene</scene>) | ||
Revision as of 03:48, 5 July 2010
The glutamate receptor is the ion channel that keeps neurons in touch. Several studies have revealed the structures of portions of this receptor [1][2][3][4]. Groundbreaking work elucidated the structure of a complete functional glutamate receptor[5][6] and that structure, 3kg2, is the subject of this page.
Contents |
Structure of the Glutamate Receptor (GluA2)
The homomeric rat GluA2 receptor arranged in a 'Y'-shape with the [7]. .
| |||||||
| glutamate receptor (3kg2), resolution 3.6Å () | |||||||
|---|---|---|---|---|---|---|---|
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
Domains
The subunits themselves are modular [8]and the major domains are found in layers in the tetrameric structure.
- The 'top' layer is composed of the
- participates directly in agonist/competitive antagonist binding, affects activation gating, and is the portion that forms the 'middle' layer.
- in the structure.
- [9], was studied as a treatment for stroke because it had demonstrated neuroprotective efficacy in experimental models of stroke and tolerability in healthy volunteers; however, in a multicenter, double-blind, randomized, placebo-controlled phase II trial, it was found to have significant sedative effects in patients with acute stroke which precludes its further development as a neuroprotective agent[10].
- is the portion that forms the membrane-spanning on the 'bottom' of the solved structure.
- To help give a better idea of how the glutamate receptor is oriented on the cell surface in the membrane lipid bilayer, as calculated by the Orientations of Proteins in Membranes database (University of Michigan, USA) is shown with the red patch of spheres indicating the boundary of the hydrophobic core closet to the outside of the cell and the dark blue patch of spheres indicating the boundary closest to the inside of the cell.

- The carboxy-terminal domain that plays a role in both receptor localization and regulation is not seen in the structure but would be below the transmembrane domain as it is cytoplasmic.
Transmembrane domain architecture
I NEED TO LABEL THE SEGMENTS BY COLOR- SHOULD I LIST THEM TOO? PROBABLY!!
Segments shown
To better show the contributions of each of the membrane segments to the interactions, . [Note: this scene generates a substantial surface which may take about a minute to calculate. Be patient.]
PDB Entry
3kg2 is a 4 chains structure of sequences from Rattus norvegicus. Full crystallographic information is available from OCA. Although it is billed as the first structure of a full-length glutamate receptor, the carboxy-terminal domain is not present in the structure.
Reference for the structure
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
Related Structures
- 3kgc GluA2 ligand-binding core complex bound with glutamate
- 2a5t GluN1-GluN2A ligand-binding domain heterodimer
- 2a5s GluN2A ligand-binding domain bound with glutamate
- 3h5w and 3h5v Crystal structure of the GluR2 amino-terminal domain[11]
- 1gr2 Structure of a glutamate-receptor ligand-binding core in complex with kainate[12]
- 3jpy and 3jpw Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit[13]
- 3hgh and 3hgh The N-terminal domain of a GluR6-subtype glutamate receptor[14]
See Also
References
- ↑ Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009 Jun 17;28(12):1812-23. Epub 2009 May 21. PMID:19461580 doi:10.1038/emboj.2009.140
- ↑ Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009 Jun;16(6):631-8. Epub 2009 May 24. PMID:19465914 doi:10.1038/nsmb.1613
- ↑ Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009 Dec 16;28(24):3910-20. Epub . PMID:19910922 doi:10.1038/emboj.2009.338
- ↑ Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998 Oct 29;395(6705):913-7. PMID:9804426 doi:10.1038/27692
- ↑ Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
- ↑ Wollmuth LP, Traynelis SF. Neuroscience: Excitatory view of a receptor. Nature. 2009 Dec 10;462(7274):729-31. PMID:20010675 doi:10.1038/462729a
- ↑ Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
- ↑ Wo ZG, Oswald RE. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 1995 Apr;18(4):161-8. PMID:7539962
- ↑ Turski L, Huth A, Sheardown M, McDonald F, Neuhaus R, Schneider HH, Dirnagl U, Wiegand F, Jacobsen P, Ottow E. ZK200775: a phosphonate quinoxalinedione AMPA antagonist for neuroprotection in stroke and trauma. Proc Natl Acad Sci U S A. 1998 Sep 1;95(18):10960-5. PMID:9724812
- ↑ Walters MR, Kaste M, Lees KR, Diener HC, Hommel M, De Keyser J, Steiner H, Versavel M. The AMPA antagonist ZK 200775 in patients with acute ischaemic stroke: a double-blind, multicentre, placebo-controlled safety and tolerability study. Cerebrovasc Dis. 2005;20(5):304-9. Epub 2005 Aug 30. PMID:16131799 doi:10.1159/000087929
- ↑ Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009 Jun 17;28(12):1812-23. Epub 2009 May 21. PMID:19461580 doi:10.1038/emboj.2009.140
- ↑ Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998 Oct 29;395(6705):913-7. PMID:9804426 doi:10.1038/27692
- ↑ Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009 Dec 16;28(24):3910-20. Epub . PMID:19910922 doi:10.1038/emboj.2009.338
- ↑ Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009 Jun;16(6):631-8. Epub 2009 May 24. PMID:19465914 doi:10.1038/nsmb.1613
Page started with original page seeded by OCA on Wed Dec 16 11:24:54 2009 for 3kg2.