Aminopeptidase
From Proteopedia
| Line 1: | Line 1: | ||
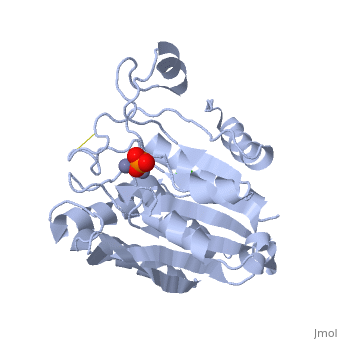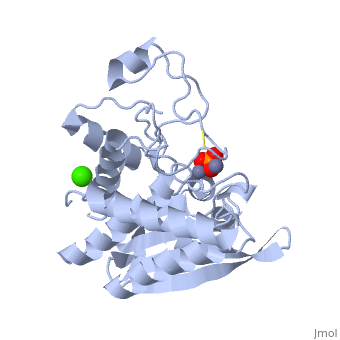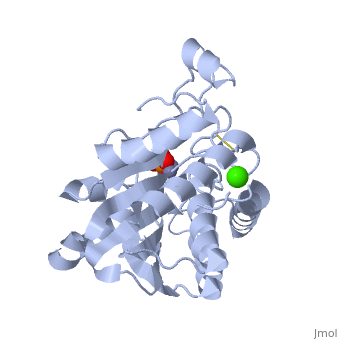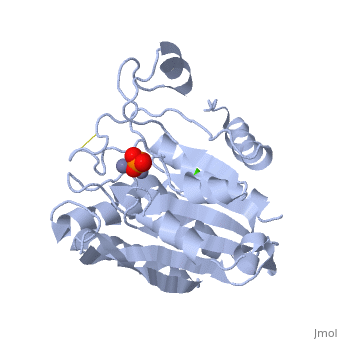
| + | [[Image:1xjo_cartoon.png|left|200px|thumb|Crystal Structure of Aminopeptidase [[1xjo]]]] | ||
| + | {{STRUCTURE_1xjo| PDB=1xjo | SIZE=300| SCENE= |right|CAPTION=Aminopeptidase [[1xjo]] }} | ||
| + | |||
'''Aminopeptidases''' catalyze a release of an N-terminal amino acid from a peptide, amide, or arylamide. | '''Aminopeptidases''' catalyze a release of an N-terminal amino acid from a peptide, amide, or arylamide. | ||
Revision as of 11:34, 30 December 2010
Aminopeptidases catalyze a release of an N-terminal amino acid from a peptide, amide, or arylamide.
S. griseus aminopeptidase
|
S. griseus aminopeptidase (SGAP) cleaves the N-terminal amino acid from a peptide or protein, and is specific for larger hydrophobic acids, especially leucine. No cleavage occurs if the next residue is proline.
The of the enzyme contains two Zn2+ ions with His85 and Asp160 as ligands for one ion, and Glu132 and His247 as ligands for the second ion. Asp97 is a common ligand to both ions. What appears to be a phosphate anion is bound to both zinc atoms, replacing the water molecule/hydroxide ion normally found in this class of enzyme.
Additional Resources
For additional information, see: Amino Acid Synthesis & Metabolism
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Joel L. Sussman, Eran Hodis