Immunodeficiency virus protease
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='2nmz' size='500' side='right' background='none' scene='User:David_Canner/Sandbox_HIV/Opening/2' caption='Structure of HIV Protease'> | |
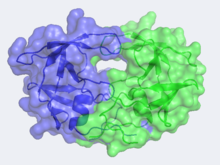
| - | HIV is | + | [[Image:CannergreyHIV2.png|220px|left]][[Human Immunodeficiency Virus]] (HIV) is the cause of Acquired Immunodeficiency Syndrome (AIDS). HIV directs the synthesis of several polyproteins, which each consist of several tandemly linked proteins. The maturation of the virus to its infectious form requires that these polyproteins be cleaved to their component proteins. <scene name='User:David_Canner/Sandbox_HIV/Opening/2'>HIV-1 protease</scene>, a homodimeric enzyme, is responsible for doing so and is therefore crucial to the virus's infectious capacity. |
| - | HIV-1 | + | ===Structure of HIV-1 Protease=== |
| + | The X-ray structure of HIV-1 protease reveals that it is composed of <scene name='User:David_Canner/Sandbox_HIV/Identical_subunits/1'>two symmetrically related subunits</scene>, each consisting of 99 amino acid residues. The subunits come together in such as way as to <scene name='User:David_Canner/Sandbox_HIV/Tunnel/1'>form a tunnel where they meet</scene>. This tunnel is of critical importance because the active site of the protease is located in its interior. The active site consists of <scene name='User:David_Canner/Sandbox_HIV/Catalytic_triad/2'> two Asp-Thr-Gly catalytic triads</scene>, making it a member of the aspartyl protease family. The two Asp's are <scene name='User:David_Canner/Sandbox_HIV/Catalytic_asp/1'>essential catalytic residues</scene> that activate a water molecule to hydrolytically cleave the polyprotein that binds in the tunnel.<ref>PMID:1799632</ref> You may be wondering how a polyprotein makes its way into the active-site tunnel, as the<scene name='User:David_Canner/Sandbox_HIV/Narrow_tunnel/1'> tunnel appears to be too narrow </scene> to admit it. The key is the two flexible flaps on the top of the tunnel that <scene name='User:David_Canner/Sandbox_HIV/Hiv_tunnel_morph/3'>move to allow proteins </scene>to enter the tunnel. The flaps <scene name='User:David_Canner/Sandbox_HIV/Hiv_tunnel_morph_flaps/2'>undergo a dramatic movement</scene>, shifting from an open to a closed conformation to bind the target in an appropriate conformation for cleavage. | ||
| - | + | ===Medical Implications=== | |
| - | + | There currently is no cure or vaccine against HIV. Researchers, however, have discovered treatments that can halt and even reverse the progression of AIDS, due in large part to our understanding of the structure of HIV-1 protease. <scene name='User:David_Canner/Sandbox_HIV/Saquinavir/4'>Saquinavir</scene> was the first protease inhibitor approved by the FDA for the treatment of HIV. It inhibits HIV protease by <scene name='User:David_Canner/Sandbox_HIV/Saquinavir_tunnel/1'>binding tightly in the active site tunnel</scene>, preventing the binding of polyproteins. Its chemical structure mimics the tetrahedral intermediate of the hydrolytic reaction, thereby <scene name='User:David_Canner/Sandbox_HIV/Saquinavir_cat/1'>interacting strongly with the catalytic triad</scene>.<ref>PMID:17243183</ref> Saquinavir is essentially an uncleavable ligand, as indicated by the <scene name='User:David_Canner/Sandbox_HIV/Hiv_morph2/9'> similar conformational changes in the protease flaps </scene> on binding saquinavir or a polypeptide . Other drugs used to treat HIV infection that inhibit <scene name='User:David_Canner/Sandbox_HIV/Inhibitor_intro/1'>HIV protease</scene> include <scene name='User:David_Canner/Sandbox_HIV/Indinavir/2'>Indinavir </scene> ([[1hsg]]), <scene name='User:David_Canner/Sandbox_HIV/Ritonavir/1'>Ritonavir</scene> ([[1hxw]]), and <scene name='User:David_Canner/Sandbox_HIV/Nelfinavir/2'>Nelfinavir</scene> ([[1ohr]]). | |
| - | + | __NOEDITSECTION__ | |
| - | + | __NOTOC__ | |
| - | Other drugs used to treat | + | </StructureSection> |
| - | + | ||
| - | + | ||
==Additional Resources== | ==Additional Resources== | ||
| Line 17: | Line 16: | ||
==References== | ==References== | ||
| - | + | <references/> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 08:06, 1 December 2010
| |||||||||||
Additional Resources
For additional information, see: Human Immunodeficiency Virus
References
- ↑ Spinelli S, Liu QZ, Alzari PM, Hirel PH, Poljak RJ. The three-dimensional structure of the aspartyl protease from the HIV-1 isolate BRU. Biochimie. 1991 Nov;73(11):1391-6. PMID:1799632
- ↑ Tie Y, Kovalevsky AY, Boross P, Wang YF, Ghosh AK, Tozser J, Harrison RW, Weber IT. Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir. Proteins. 2007 Apr 1;67(1):232-42. PMID:17243183 doi:10.1002/prot.21304
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Michal Harel, Eran Hodis, Mark Hoelzer, Marius Mihasan, David Canner, Eric Martz, Ann Taylor, Wayne Decatur, Alexander Berchansky, Jaime Prilusky, Karsten Theis