Aldolase
From Proteopedia
| Line 34: | Line 34: | ||
The regulation of fructose 1,6-bisphosphate aldolase is not well understood, but the understanding is every-increasing. As it is currently observed, aldolase C appears to be regulated mainly by the gene expression--the concentration of mRNA in the cytoplasm.<ref>Paolella, G, Buono, P, Mancini, F P, Izzo, P, and Salvatore, F. "Structure and expression of mouse aldolase genes." Eur. J. Biochem.. 156. (1986): 229-235.</ref> It is also known that adenosine 3',5'-cyclicmonophosphate (cAMP) affects the expression of the gene. cAMP concentration has been positively correlated with aldolase C expression. It is believed that cAMP acts upon a section of the promotor region, distal element D, causing the transcriptional promoter, NGFI-B, to bind. Once bound, the promoter activates the transcription of the gene coding for fructose bisphosphate aldolase.<ref>Buono, P, Cassano, S, Alfieri, A, Mancini, A, and Salvatore, F. "Human aldolase C gene expression is regulated by adenosine 30,50-cyclic monophosphate (cAMP) in PC12 cells." Gene. 291. (2002): 115-121.</ref> Given the inhibitory effects of an oxidant in the presence of aldolase, it is possible that this could be a mechanism of regulation of the enzyme. The deactivation that accompanies the oxidation of the surface thiol of Cys72 could be used intracellularly to slow the catalysis of the enzyme and regulate glycolysis.<ref name="kinetics" /> | The regulation of fructose 1,6-bisphosphate aldolase is not well understood, but the understanding is every-increasing. As it is currently observed, aldolase C appears to be regulated mainly by the gene expression--the concentration of mRNA in the cytoplasm.<ref>Paolella, G, Buono, P, Mancini, F P, Izzo, P, and Salvatore, F. "Structure and expression of mouse aldolase genes." Eur. J. Biochem.. 156. (1986): 229-235.</ref> It is also known that adenosine 3',5'-cyclicmonophosphate (cAMP) affects the expression of the gene. cAMP concentration has been positively correlated with aldolase C expression. It is believed that cAMP acts upon a section of the promotor region, distal element D, causing the transcriptional promoter, NGFI-B, to bind. Once bound, the promoter activates the transcription of the gene coding for fructose bisphosphate aldolase.<ref>Buono, P, Cassano, S, Alfieri, A, Mancini, A, and Salvatore, F. "Human aldolase C gene expression is regulated by adenosine 30,50-cyclic monophosphate (cAMP) in PC12 cells." Gene. 291. (2002): 115-121.</ref> Given the inhibitory effects of an oxidant in the presence of aldolase, it is possible that this could be a mechanism of regulation of the enzyme. The deactivation that accompanies the oxidation of the surface thiol of Cys72 could be used intracellularly to slow the catalysis of the enzyme and regulate glycolysis.<ref name="kinetics" /> | ||
| + | |||
| + | ==3D structures of Aldolase== | ||
| + | |||
| + | ===Fructose–1,6-bisphosphate aldolase=== | ||
| + | |||
| + | [[1ojx]], [[1ok6]] – TptFBPA – Thermoproteus tenax | ||
| + | |||
| + | [[3qrh]] – EncFBPA – Encephalitozoon cuniculi | ||
| + | |||
| + | [[3qm3]] - FBPA – Campylobacter jejuni | ||
| + | |||
| + | [[3q94]] – FBPA – Bacullus anthracis | ||
| + | |||
| + | [[3c4u]]– HpFBPA – Helicobacter pylori | ||
| + | |||
| + | [[1zah]], [[1fdj]], [[1ewd]], [[1ewe]], [[1ex5]], [[1ado]] - rFBPA – rabbit | ||
| + | |||
| + | [[3dfn]], [[3dfp]], [[3dfq]], [[3dft]], [[2bv4]], [[3b8d]] – rFBPA (mutant) | ||
| + | |||
| + | [[3kx6]] – FBPA – Babesia bovis | ||
| + | |||
| + | [[3gak]] – GiFBPA – Giardia intestinalis | ||
| + | |||
| + | [[3ekl]], [[3ekz]] – MtFBPA – Mycobacterium tuberculosis | ||
| + | |||
| + | [[2qap]], [[1epx]] – LmFBPA – Leishmania mexicana | ||
| + | |||
| + | [[1a5c]] – PfFBPA – Plasmodium falciparum | ||
| + | |||
| + | [[2iqt]] – FBPA – Porphyromonas gingivalis | ||
| + | |||
| + | [[2fjk]] – FBPA – Thermus caldophilus | ||
| + | |||
| + | [[1xfb]], [[1qo5]], [[2ald]], [[1ald]] – hFBPA – human | ||
| + | |||
| + | [[1xdl]], [[1xdm]] – hFBPA (mutant) | ||
| + | |||
| + | [[1gyn]], [[1l6w]], [[1dos]], [[1zen]] – EcFBPA - Escherichia coli | ||
| + | |||
| + | [[1f2j]] – FBPA – Trypanosoma brucei | ||
| + | |||
| + | [[1fba]] – FBPA – Drosophila melanogaster | ||
| + | |||
| + | FBPA binary complex | ||
| + | |||
| + | [[2yce]], [[1ok4]] – TptFBPA + reaction intermediate | ||
| + | |||
| + | [[1w8s]] – TptFBPA + FBP | ||
| + | |||
| + | [[3mbd]] – EncFBPA + phosphate | ||
| + | |||
| + | [[3mbf]] - EncFBPA + FBP | ||
| + | |||
| + | [[3n9r]], [[3n9s]], [[3c52]], [[3c56]] - HpFBPA + inhibitor | ||
| + | |||
| + | [[3mmt]] – FBPA + FBP – Bartonella henselae | ||
| + | |||
| + | [[1zai]] - rFBPA + FBP | ||
| + | |||
| + | [[6ald]] - rFBPA (mutant) + FBP | ||
| + | |||
| + | [[2quv]] - rFBPA + phosphate | ||
| + | |||
| + | [[2qut]] - rFBPA + reaction intermediate | ||
| + | |||
| + | [[3dfo]], [[3dfs]], [[2quu]] - rFBPA (mutant) + reaction intermediate | ||
| + | |||
| + | [[1j4e]] - rFBPA (mutant) + substrate | ||
| + | |||
| + | [[2ot0]] – rFBPA + Wiskott-Aldrich syndrome protein C-terminal | ||
| + | |||
| + | [[2ot1]], [[1zaj]], [[1zal]] – rFBPA + inhibitor | ||
| + | |||
| + | [[3lge]] – rFBPA + Sorting Nexin-9 | ||
| + | |||
| + | [[3gay]] – GiFBPA + inhibitor | ||
| + | |||
| + | [[3gb6]] – GiFBPA + FBP | ||
| + | |||
| + | [[2isv]], [[2isw]] - GiFBPA + oxamate | ||
| + | |||
| + | [[3elf]] – MtFBPA + FBP | ||
| + | |||
| + | [[2qdg]] - LmFBPA + FBP | ||
| + | |||
| + | [[2qdh]] – LmFBPA + inhibitor | ||
| + | |||
| + | [[2eph]], [[2pc4]] – PfFBPA + BPTRAP C-terminal | ||
| + | |||
| + | [[4ald]] – hFBPA + FBP | ||
| + | |||
| + | [[1rv8]], [[1rvg]] – FBPA + metal – Thermus aquaticus | ||
| + | |||
| + | [[1b57]] – EcFBPA + oxamate | ||
| + | |||
| + | ===Tagatose–1,6-bisphosphate aldolase=== | ||
| + | |||
| + | [[3myo]], [[3myp]], [[3mhf]]– SpTBPA – Streptococcus pyogenes | ||
| + | |||
| + | [[3mhg]] - SpTBPA + reaction intermediate | ||
| + | |||
| + | [[3kao]] – SaTBPA – Staphylococcus aureus | ||
| + | |||
| + | [[1gvf]] - EcTBPA | ||
| + | |||
| + | ===Fuculose–1-phosphate aldolase=== | ||
| + | |||
| + | [[2opi]] – FPA – Bacteroides thetaiotaomicron | ||
| + | |||
| + | [[2flf]], [[2fk5]] – TtFPA]] - Thermus thermophilus | ||
| + | |||
| + | [[1fua]], [[2fua]], [[3fua]] – EcFPA | ||
| + | |||
| + | [[1e46]], [[1e47]], [[1e48]], [[1e49]], [[1e4a]], [[1e4b]], [[1e4c]], [[1dzu]], [[1dzw]], [[1dzx]], [[1dzy]], [[1dzz]] – EcFPA (mutant) | ||
| + | |||
| + | [[4fua]] – EcFPA + oxamate | ||
| + | |||
| + | ===Deoxyribose-phosphate aldolase=== | ||
| + | |||
| + | [[3r12]], [[3r13]], [[1pvt]], [[1o0y]] – TmDERA – Thermotoga maritime | ||
| + | |||
| + | [[3ndo]] – MsDERA – Mycobacterium smegmatis | ||
| + | |||
| + | [[3ng3]] – MsDERA + aldehyde | ||
| + | |||
| + | [[2a4a]] – DERA – Plasmodium yoelii | ||
| + | |||
| + | [[1vcv]] – DERA – Pyrobaculum aerophilum | ||
| + | |||
| + | [[1p1x]] – EcDERA | ||
| + | |||
| + | [[1jcj]], [[1jcl]] – EcDERA (mutant) + reaction intermediate | ||
| + | |||
| + | [[1n7k]] – DERA – Aeropyrum pernix | ||
| + | |||
| + | [[1mzh]] – AaDERA]] - Aquifex aeolicus | ||
| + | |||
| + | ===Dehydroneopterin aldolase=== | ||
| + | |||
| + | [[3r2e]] – DHNPA – Yersinia pestis | ||
| + | |||
| + | [[2o90]] – EcDHNPA + neopterin | ||
| + | |||
| + | [[2nm2]], [[2nm3]], [[1u68]], [[2dhn]] - SaDHNPA + neopterin | ||
| + | |||
| + | [[1rri]], [[1rrw]], [[1rry]], [[1rs2]], [[1rs4]], [[1rsd]], [[1rsi]], [[1dhn]] - SaDHNPA + inhibitor | ||
| + | |||
| + | [[1z9w]] – MtDHNPA | ||
| + | |||
| + | [[1sql]] – DHNPA + guanine – Arabidopsis thaliana | ||
| + | |||
| + | ===HPCH/HPAI aldolase=== | ||
| + | |||
| + | [[3qz6]] – HPA – Desulfitobacterium hafniense | ||
| + | |||
| + | [[2v5j]] – EcHPA | ||
| + | |||
| + | [[2v5k]] – EcHPA + oxamate | ||
| + | |||
| + | ===Sialic acid aldolase=== | ||
| + | |||
| + | [[3lbm]] – EcSAA | ||
| + | |||
| + | [[3lcf]], [[3lcg]], [[3lch]], [[3lci]], [[3lcl]], [[2wnq]], [[2wo5]] - EcSAA (mutant) | ||
| + | |||
| + | [[3lbc]] – EcSAA + L-arabinose | ||
| + | |||
| + | [[2wnn]] – EcSAA + pyruvate <br /> | ||
| + | [[2wnz]], [[2wkj]] - EcSAA (mutant) + pyruvate <br /> | ||
| + | [[2wpb]] - EcSAA (mutant) + pyruvate + inhibitor | ||
| + | |||
| + | [[3lcx]] - EcSAA L-KDO (mutant) | ||
| + | |||
| + | [[3lcw]] - EcSAA L-KDO (mutant) + hydroxypyruvate <br /> | ||
| + | |||
| + | ===Oxoadipate aldolase=== | ||
| + | |||
| + | [[3noj]] – PpCHA-ALD – Pseudomonas putida | ||
| + | |||
| + | ===Oxovalerate aldolase=== | ||
| + | |||
| + | [[1nvm]] – OVA + acetaldehyde dehydrogenase]] - Pseudomonas | ||
| + | |||
| + | ===Deoxydephosphogluconate aldolase=== | ||
| + | |||
| + | [[3nzr]] – DDPGA – Vibrio fischeri | ||
| + | |||
| + | [[2r91]], [[2r94]] – TptDDPGA | ||
| + | |||
| + | [[2yw3]], [[2yw4]] – TtDDPGA | ||
| + | |||
| + | [[2nuw]], [[2nux]] – SaDDPGA – Sulfolobus acidocaldarius | ||
| + | |||
| + | [[1vlw]] – TmDDPGA | ||
| + | |||
| + | [[1w37]] - SsDDPGA – Sulfolobus solfataricus | ||
| + | |||
| + | [[1mxs]], [[1kga]] – PpDDPGA | ||
| + | |||
| + | [[1eun]], [[1fq0]] – EcDDPGA | ||
| + | |||
| + | [[1fwr]] - EcDDPGA (mutant) | ||
| + | |||
| + | DDPGA complex | ||
| + | |||
| + | [[1nuy]] – SaDDPGA + pyruvate <br /> | ||
| + | [[1wa3]] - TmDDPGA + pyruvate <br /> | ||
| + | [[1w3i]] - SsDDPGA + pyruvate <br /> | ||
| + | [[1w3n]] - SsDDPGA + gluconate | ||
| + | |||
| + | [[1w3t]] - SsDDPGA + gluconate + pyruvate <br /> | ||
| + | [[1eua]] - EcDDPGA + pyruvate <br /> | ||
| + | |||
| + | ===Deoxydephosphooctonate aldolase=== | ||
| + | |||
| + | [[3e0i]], [[2nxi]], [[1fx6]] – AaDDPOA | ||
| + | |||
| + | [[2ef9]], [[2nws]], [[2nx1]], [[2nx3]], [[2nxg]], [[2nxh]], [[1t99]] – AaDDPOA (mutant) | ||
| + | |||
| + | [[1x8f]] – EcDDPOA | ||
| + | |||
| + | [[3fs2]] – DDPOA – Bruciella melitensis | ||
| + | |||
| + | [[3e9a]] – DDPOA – Vibrio cholerae | ||
| + | |||
| + | [[2qkf]] – DDPOA – Neisseria meningitides | ||
| + | |||
| + | DDPOA binary complex | ||
| + | |||
| + | [[1fxp]] – AaDDPOA + Cd | ||
| + | |||
| + | [[1pck]], [[1fwn]], [[1fws]] - AaDDPOA + PEP | ||
| + | |||
| + | [[1pcw]], [[1pe1]], [[1jcx]] - AaDDPOA + inhibitor | ||
| + | |||
| + | [[1lrn]] - AaDDPOA (mutant) + Cd | ||
| + | |||
| + | [[2nwr]], [[1t96]], [[1lro]] - AaDDPOA (mutant) + PEP | ||
| + | |||
| + | [[3e12]] – AaDDPOA + KDO8P | ||
| + | |||
| + | [[1x6u]] - EcDDPOA + KDO8P | ||
| + | |||
| + | [[1q3n]] - EcDDPOA + PEP | ||
| + | |||
| + | [[1phq]], [[1phw]], [[1pl9]] - EcDDPOA + substrate analog | ||
| + | |||
| + | DDPOA tertiary complex | ||
| + | |||
| + | [[1fy6]] - AaDDPOA + arabinose + Cd | ||
| + | |||
| + | [[1jcy]], [[1fwt]], [[1fww]], [[1fxq]] - AaDDPOA + PEP + sugar | ||
| + | |||
| + | [[2a2i]], [[1zha]], [[1zji]], [[1t8x]], [[1lrq]] - AaDDPOA (mutant) + PEP + arabinose | ||
| + | |||
| + | [[2a21]] - AaDDPOA + PEP + phosphate | ||
| + | |||
| + | ===Deoxydephosphogalactonate aldolase=== | ||
| + | |||
| + | [[2v81]] – EcKDPGAL | ||
| + | |||
| + | [[2c0a]] – EcKDPGAL (mutant) | ||
| + | |||
| + | [[2v82]] – EcKDPGAL + 2-keto-deoxy-galactose | ||
| + | |||
| + | ===Deoxydephosphoheptonate aldolase=== | ||
| + | |||
| + | [[1vr6]] – TmKDPHAL | ||
| + | |||
| + | ===Deoxygalactarate aldolase=== | ||
| + | |||
| + | [[1dxe]] – EcDGA | ||
| + | |||
| + | [[1dxf]] – EcDGA + pyruvate <br /> | ||
| + | |||
| + | ===Aldolase class II=== | ||
| + | |||
| + | [[3ocr]] – ALDII – Pseudomonas syringae | ||
| + | |||
| + | [[2vws]] – EcALDII | ||
| + | |||
| + | [[2vwt]] – EcALDII + pyruvate <br /> | ||
| + | |||
| + | ===Sphingosin-1-phosphate aldolase=== | ||
| + | |||
| + | [[3mc6]] – SCDPL1 (mutant) – yeast | ||
| + | |||
| + | ===Rhamnulose-1-phosphate aldolase=== | ||
| + | |||
| + | [[1gt7]] – EcRPA | ||
| + | |||
| + | [[2v9g]], [[2v9o]], [[2uyu]], [[2uyv]], [[2v9e]], [[2v9f]], [[2v9i]], [[2v9l]], [[2v9m]], [[2v9n]], [[2v29]], [[2v2a]], [[2v2b]], [[1ojr]] – EcRPA (mutant) | ||
| + | |||
| + | ===Oxoglutarate aldolase=== | ||
| + | |||
| + | [[3m6y]] – OGA – Bacillus cereus | ||
| + | |||
| + | [[1wau]], [[1wbh]] – EcOGA (mutant) | ||
| + | |||
| + | ===LsrF aldolase=== | ||
| + | |||
| + | [[3gkf]] – EcLsrFA | ||
| + | |||
| + | [[3glc]], [[3gnd]] – EcLsrFA + ribose derivative | ||
| + | |||
| + | ===Threonine aldolase=== | ||
| + | |||
| + | [[2fm1]], [[1m6s]], [[1jg8]] – TmThrA | ||
| + | |||
| + | [[1lw4]], [[1lw5]] – TmThrA + amino acid | ||
| + | |||
| + | [[1svv]] – ThrA – Leishmania major | ||
| + | |||
| + | ===Phenylserine aldolase=== | ||
| + | |||
| + | [[1v72 – PpFSA | ||
| + | |||
| + | [[1j2w]], [[1ub3]] - TtALD | ||
==Additional Resources== | ==Additional Resources== | ||
Revision as of 11:25, 4 May 2011
Fructose Bisphosphate Aldolase
| |||||||||
| 4ald, resolution 2.80Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Activity: | Fructose-bisphosphate aldolase, with EC number 4.1.2.13 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Introduction and Structure
Fructose bisphosphate aldolase is an enzyme in glycolysis and gluconeogenesis. Glycolyis is responsible for the conversion of glucose into two three-carbon pyruvate molecules without the need for oxygen. The process generates two net ATP. The overall reaction is:
Glucose + 2 NAD+ + 2 ADP + 2 Pi --> 2 pyruvate (3-carbon product) + 2 NADH + 2 ATP + 2 H20 + 4 H+
Gluconeogenesis is responsible for maintaining the appropriate levels of blood glucose in animals by generating glucose from non-carbohydrate precursors. Gluconeogenesis can make glucose from lactate, pyruvate, citric acid cycle intermediates and from most amino acids (the exceptions being leucine and lysine). The common intermediate for all of the precursors on their way to becoming glucose must be oxaloacetate.
The aldolase catalyzes the reversible cleavage of fructose-1,6-bisphosphate into dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP). Different isozymes of aldolase can also catalyze the cleavage of fructose 1-phosphate to diydroxyacetone and glyceraldehyde (GA). Different isozymes exhibit preferences for either or both of the substrates, depending on the role of the aldolase (i.e. gluconeogenesis versus glycolysis).[1]
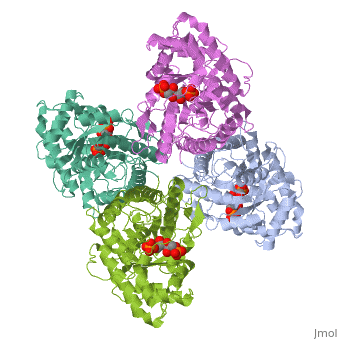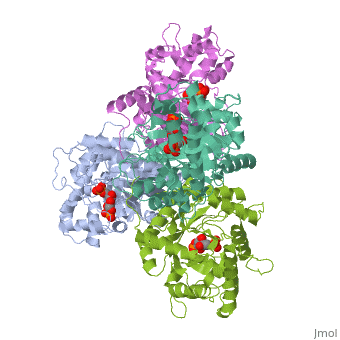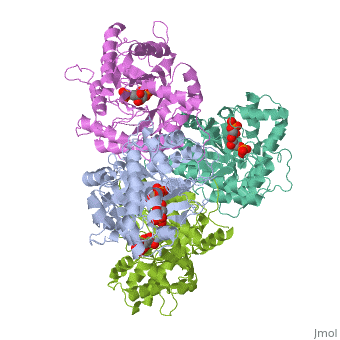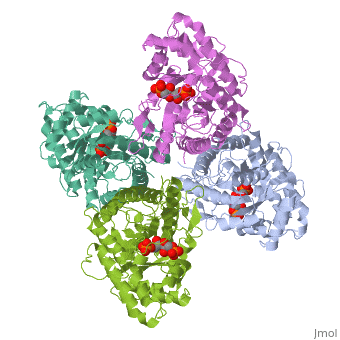
While it can exist as a monomer, it normally exists as a . The enzyme is an a/B protein with a TIM beta/alpha beta fold. The fold designation is based upon the nine alpha helices and eight parallel beta sheets in a closed barrel of each monomeric subunit. It is part of the aldolase superfamily and the class I aldolases.[2] can be seen in their specific regions mostly concentric to the active site, represented by the blue and red residues.
Although some form of fructose bisphosphate aldolase is present in nearly all living things, certain isoforms carry a large degree of conservation. The enzyme from rabbit muscle has nearly the tertiary and primary structure as the enzyme in human muscle. As a result, implications from rabbit muscle aldolase also reveal a great deal about the human forms of the enzyme. [3]
Binding and Catalysis
As an enzyme, the aldolase must not only encourage and favor the hydrolysis of fructose 1,6-bisphosphate, but also bind the substrate so as to hold it in the active site. The main-chain nitrogens of Ser271 and Gly272 hold the 1-phosphate group while the Lys41, Arg42 and Arg303 residues hold the 6-phosphate group. The five proposed binding residues are in close proximity to the catalytic Lys229, implicating them as participants in the binding process.[4] The , which sits just outside of the barrel and catalytic site, of the enzyme also appears to contribute to the catalytic process of the aldolase. Mutations or suppression of the final tyrosine residue (Tyr363) causes a notable drop in the activity of the enzyme. Two cysteine residues have also been implicated in the catalytic process. Though they do not appear to be necessary for catalysis, modification of them does result in a decrease in catalytic activity. The two Cys residues are far from the active site, but do impact the movement of the C-terminus of the enzyme, which further implicates the terminus as participatory in the catalysis.
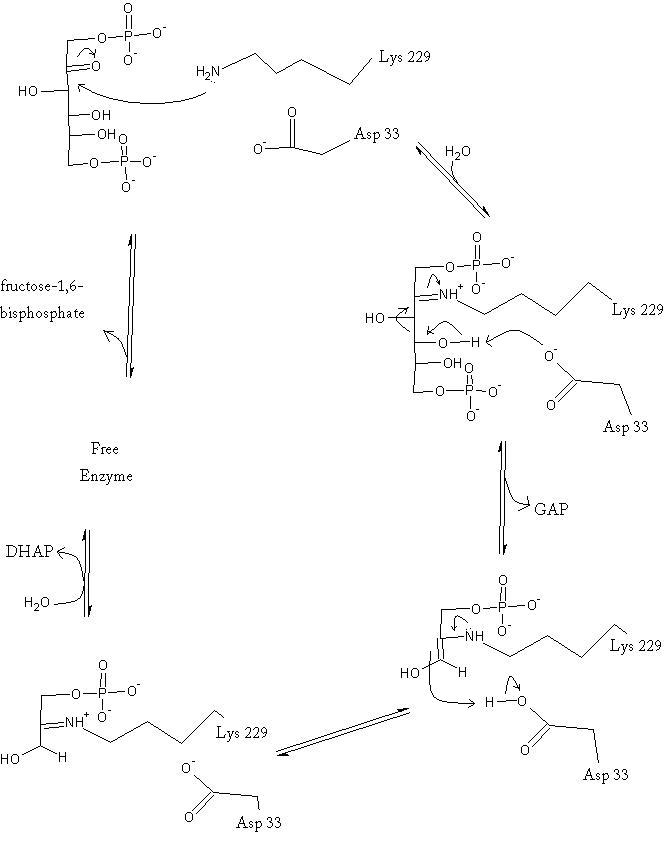
The reaction is an aldol cleavage, or otherwise termed, retro aldo condensation. Catalysis occurs first when the nucleophilic ε-amine group of Lys229 attacks the carbonyl carbon of the substrate (FBP) in its open-ring state, pushing an electron pair to the oxygen of the carbonyl. The oxygen is protonated and leaves as water as a protonated is produced (an imine resulting from a ketone and amine) with the open-ring form of FBP, accompanied by electrostatic stabilization from Aldol cleavage between C3 and C4 produces GAP and an enamine precursor to DHAP.[1] The cleavage is facilitated by the positive charge from the Schiff base. The subsequent electron movement, which alleviates the positive charge, also breaks the C3-C4 bond.[3] Tautomerization, protonation and the hydrolysis of the Schiff base produce the final product of DHAP and regenerate the enzyme. The catalysis is driven by the more favorable stability of the protonated Schiff base compared to the enolate that would appear in basic catalysis pathways.[1]
Kinetics
Isotopic labelling has revealed the rate-determining step for the reaction. Either the carbon-carbon bond cleavage or the release of glyceraldehyde-3-phosphate comprise the slow step of the catalysis reaction; however, studies do indicate that the GAP release is likely the slowest step.[3]
It has been shown that aldolase is inhibited allosterically by oxidized glutathione, which is an oxidizing species biologically present. The glutathione oxidizes a thiol 25 angstroms from the catalytic site, which subsequently causes a drop in catalytic activity. In addition, the enzyme shows no positive cooperativity, despite being an oligomer. In fact, kinetics data actually show that the enzyme exhibits negative cooperativity. Thus the catalysis is highly compartmentalized within each subunit and binding causes little distal change of the enzymes structure.[5]
Regulation
The regulation of fructose 1,6-bisphosphate aldolase is not well understood, but the understanding is every-increasing. As it is currently observed, aldolase C appears to be regulated mainly by the gene expression--the concentration of mRNA in the cytoplasm.[6] It is also known that adenosine 3',5'-cyclicmonophosphate (cAMP) affects the expression of the gene. cAMP concentration has been positively correlated with aldolase C expression. It is believed that cAMP acts upon a section of the promotor region, distal element D, causing the transcriptional promoter, NGFI-B, to bind. Once bound, the promoter activates the transcription of the gene coding for fructose bisphosphate aldolase.[7] Given the inhibitory effects of an oxidant in the presence of aldolase, it is possible that this could be a mechanism of regulation of the enzyme. The deactivation that accompanies the oxidation of the surface thiol of Cys72 could be used intracellularly to slow the catalysis of the enzyme and regulate glycolysis.[5]
3D structures of Aldolase
Fructose–1,6-bisphosphate aldolase
1ojx, 1ok6 – TptFBPA – Thermoproteus tenax
3qrh – EncFBPA – Encephalitozoon cuniculi
3qm3 - FBPA – Campylobacter jejuni
3q94 – FBPA – Bacullus anthracis
3c4u– HpFBPA – Helicobacter pylori
1zah, 1fdj, 1ewd, 1ewe, 1ex5, 1ado - rFBPA – rabbit
3dfn, 3dfp, 3dfq, 3dft, 2bv4, 3b8d – rFBPA (mutant)
3kx6 – FBPA – Babesia bovis
3gak – GiFBPA – Giardia intestinalis
3ekl, 3ekz – MtFBPA – Mycobacterium tuberculosis
2qap, 1epx – LmFBPA – Leishmania mexicana
1a5c – PfFBPA – Plasmodium falciparum
2iqt – FBPA – Porphyromonas gingivalis
2fjk – FBPA – Thermus caldophilus
1xfb, 1qo5, 2ald, 1ald – hFBPA – human
1gyn, 1l6w, 1dos, 1zen – EcFBPA - Escherichia coli
1f2j – FBPA – Trypanosoma brucei
1fba – FBPA – Drosophila melanogaster
FBPA binary complex
2yce, 1ok4 – TptFBPA + reaction intermediate
1w8s – TptFBPA + FBP
3mbd – EncFBPA + phosphate
3mbf - EncFBPA + FBP
3n9r, 3n9s, 3c52, 3c56 - HpFBPA + inhibitor
3mmt – FBPA + FBP – Bartonella henselae
1zai - rFBPA + FBP
6ald - rFBPA (mutant) + FBP
2quv - rFBPA + phosphate
2qut - rFBPA + reaction intermediate
3dfo, 3dfs, 2quu - rFBPA (mutant) + reaction intermediate
1j4e - rFBPA (mutant) + substrate
2ot0 – rFBPA + Wiskott-Aldrich syndrome protein C-terminal
2ot1, 1zaj, 1zal – rFBPA + inhibitor
3lge – rFBPA + Sorting Nexin-9
3gay – GiFBPA + inhibitor
3gb6 – GiFBPA + FBP
3elf – MtFBPA + FBP
2qdg - LmFBPA + FBP
2qdh – LmFBPA + inhibitor
2eph, 2pc4 – PfFBPA + BPTRAP C-terminal
4ald – hFBPA + FBP
1rv8, 1rvg – FBPA + metal – Thermus aquaticus
1b57 – EcFBPA + oxamate
Tagatose–1,6-bisphosphate aldolase
3myo, 3myp, 3mhf– SpTBPA – Streptococcus pyogenes
3mhg - SpTBPA + reaction intermediate
3kao – SaTBPA – Staphylococcus aureus
1gvf - EcTBPA
Fuculose–1-phosphate aldolase
2opi – FPA – Bacteroides thetaiotaomicron
2flf, 2fk5 – TtFPA]] - Thermus thermophilus
1e46, 1e47, 1e48, 1e49, 1e4a, 1e4b, 1e4c, 1dzu, 1dzw, 1dzx, 1dzy, 1dzz – EcFPA (mutant)
4fua – EcFPA + oxamate
Deoxyribose-phosphate aldolase
3r12, 3r13, 1pvt, 1o0y – TmDERA – Thermotoga maritime
3ndo – MsDERA – Mycobacterium smegmatis
3ng3 – MsDERA + aldehyde
2a4a – DERA – Plasmodium yoelii
1vcv – DERA – Pyrobaculum aerophilum
1p1x – EcDERA
1jcj, 1jcl – EcDERA (mutant) + reaction intermediate
1n7k – DERA – Aeropyrum pernix
1mzh – AaDERA]] - Aquifex aeolicus
Dehydroneopterin aldolase
3r2e – DHNPA – Yersinia pestis
2o90 – EcDHNPA + neopterin
2nm2, 2nm3, 1u68, 2dhn - SaDHNPA + neopterin
1rri, 1rrw, 1rry, 1rs2, 1rs4, 1rsd, 1rsi, 1dhn - SaDHNPA + inhibitor
1z9w – MtDHNPA
1sql – DHNPA + guanine – Arabidopsis thaliana
HPCH/HPAI aldolase
3qz6 – HPA – Desulfitobacterium hafniense
2v5j – EcHPA
2v5k – EcHPA + oxamate
Sialic acid aldolase
3lbm – EcSAA
3lcf, 3lcg, 3lch, 3lci, 3lcl, 2wnq, 2wo5 - EcSAA (mutant)
3lbc – EcSAA + L-arabinose
2wnn – EcSAA + pyruvate
2wnz, 2wkj - EcSAA (mutant) + pyruvate
2wpb - EcSAA (mutant) + pyruvate + inhibitor
3lcx - EcSAA L-KDO (mutant)
3lcw - EcSAA L-KDO (mutant) + hydroxypyruvate
Oxoadipate aldolase
3noj – PpCHA-ALD – Pseudomonas putida
Oxovalerate aldolase
1nvm – OVA + acetaldehyde dehydrogenase]] - Pseudomonas
Deoxydephosphogluconate aldolase
3nzr – DDPGA – Vibrio fischeri
2nuw, 2nux – SaDDPGA – Sulfolobus acidocaldarius
1vlw – TmDDPGA
1w37 - SsDDPGA – Sulfolobus solfataricus
1fwr - EcDDPGA (mutant)
DDPGA complex
1nuy – SaDDPGA + pyruvate
1wa3 - TmDDPGA + pyruvate
1w3i - SsDDPGA + pyruvate
1w3n - SsDDPGA + gluconate
1w3t - SsDDPGA + gluconate + pyruvate
1eua - EcDDPGA + pyruvate
Deoxydephosphooctonate aldolase
2ef9, 2nws, 2nx1, 2nx3, 2nxg, 2nxh, 1t99 – AaDDPOA (mutant)
1x8f – EcDDPOA
3fs2 – DDPOA – Bruciella melitensis
3e9a – DDPOA – Vibrio cholerae
2qkf – DDPOA – Neisseria meningitides
DDPOA binary complex
1fxp – AaDDPOA + Cd
1pck, 1fwn, 1fws - AaDDPOA + PEP
1pcw, 1pe1, 1jcx - AaDDPOA + inhibitor
1lrn - AaDDPOA (mutant) + Cd
2nwr, 1t96, 1lro - AaDDPOA (mutant) + PEP
3e12 – AaDDPOA + KDO8P
1x6u - EcDDPOA + KDO8P
1q3n - EcDDPOA + PEP
1phq, 1phw, 1pl9 - EcDDPOA + substrate analog
DDPOA tertiary complex
1fy6 - AaDDPOA + arabinose + Cd
1jcy, 1fwt, 1fww, 1fxq - AaDDPOA + PEP + sugar
2a2i, 1zha, 1zji, 1t8x, 1lrq - AaDDPOA (mutant) + PEP + arabinose
2a21 - AaDDPOA + PEP + phosphate
Deoxydephosphogalactonate aldolase
2v81 – EcKDPGAL
2c0a – EcKDPGAL (mutant)
2v82 – EcKDPGAL + 2-keto-deoxy-galactose
Deoxydephosphoheptonate aldolase
1vr6 – TmKDPHAL
Deoxygalactarate aldolase
1dxe – EcDGA
1dxf – EcDGA + pyruvate
Aldolase class II
3ocr – ALDII – Pseudomonas syringae
2vws – EcALDII
2vwt – EcALDII + pyruvate
Sphingosin-1-phosphate aldolase
3mc6 – SCDPL1 (mutant) – yeast
Rhamnulose-1-phosphate aldolase
1gt7 – EcRPA
2v9g, 2v9o, 2uyu, 2uyv, 2v9e, 2v9f, 2v9i, 2v9l, 2v9m, 2v9n, 2v29, 2v2a, 2v2b, 1ojr – EcRPA (mutant)
Oxoglutarate aldolase
3m6y – OGA – Bacillus cereus
LsrF aldolase
3gkf – EcLsrFA
3glc, 3gnd – EcLsrFA + ribose derivative
Threonine aldolase
1lw4, 1lw5 – TmThrA + amino acid
1svv – ThrA – Leishmania major
Phenylserine aldolase
[[1v72 – PpFSA
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ 1.0 1.1 1.2 Voet, D, Voet, J, & Pratt, C. (2008). Fundamentals of biochemistry, third edition. Hoboken, NJ: Wiley & Sons, Inc.
- ↑ Protein: fructose-1,6-bisphosphate aldolase from human (homo sapiens), muscle isozyme. (2009). Retrieved from http://scop.mrc-lmb.cam.ac.uk
- ↑ 3.0 3.1 3.2 Gefflaut, T., B. Casimir, J. Perie, and M. Willson. "Class I Aldolases: Substrate Specificity, Mechanism, Inhibitors and Structural Aspects." Prog. Biophys. molec. Biol.. 63. (1995): 301-340.
- ↑ Dalby A, Dauter Z, Littlechild JA. Crystal structure of human muscle aldolase complexed with fructose 1,6-bisphosphate: mechanistic implications. Protein Sci. 1999 Feb;8(2):291-7. PMID:10048322
- ↑ 5.0 5.1 Sygusch, J., and Beaudry, D. "Allosteric communication in mammalian muscle aldolase." Biochem. J.. 327. (1997): 717-720.
- ↑ Paolella, G, Buono, P, Mancini, F P, Izzo, P, and Salvatore, F. "Structure and expression of mouse aldolase genes." Eur. J. Biochem.. 156. (1986): 229-235.
- ↑ Buono, P, Cassano, S, Alfieri, A, Mancini, A, and Salvatore, F. "Human aldolase C gene expression is regulated by adenosine 30,50-cyclic monophosphate (cAMP) in PC12 cells." Gene. 291. (2002): 115-121.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Sophie Mullinix, Jaime Prilusky, Austin Drake, David Canner