Luciola cruciata luciferase
From Proteopedia
m (→See Also) |
|||
| Line 1: | Line 1: | ||
| - | [[Image: | + | [[Image:2d1r.png|left|200px]] |
| - | == Introduction == | + | {{STRUCTURE_2d1r| PDB=2d1r | SCENE= }} |
| - | + | ||
| + | ==Introduction== | ||
| + | |||
| + | [[Luciferase]] is a class of enzymes producing light through the process of bioluminescence. This luciferase was extracted from [http://en.wikipedia.org/wiki/Luciola_cruciata Luciola cruciata]. In the case of fireflies, it catalyses a reaction of adenylation and then an oxydative decarboxylation, changing luciferin to oxyluciferin and thus emitting light. | ||
| + | This protein is constituted of two main domains separated by a cleft. | ||
| + | |||
| + | |||
| + | ==Chemical reaction== | ||
| + | |||
| + | The enzyme catalyses the production of light, using two different conformation to catalyze two half-reactions (in the case of firefly Luciferase). | ||
| + | |||
| + | * luciferin + ATP → luciferyl adenylate + PP<sub>i</sub> | ||
| + | |||
| + | * luciferyl adenylate + O<sub>2</sub> → <scene name='Sandbox2442/Oxyluciferin/2'>oxyluciferin</scene> + <scene name='Sandbox2442/Amp/3'>AMP</scene> + light | ||
| + | |||
| + | The first half-reaction is an adenylation, the second one is an oxydative decarboxylation. | ||
| + | |||
| + | The production of light is achieved by the conversion of chemical energy into an emission of photons, resulting from the passage of the oxyluciferin's excitation state to a ground state. | ||
| + | |||
| + | ==The structure== | ||
| + | 2D1R is a 1 chain structure of sequence from [http://en.wikipedia.org/wiki/Luciola_cruciata Luciola cruciata]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2D1R OCA]. | ||
| + | The protein is shown here bound with two products, adenosine 5'-phosphate (AMP) and oxyluciferine. | ||
| + | |||
| + | |||
| + | It weights 62 kDa and is constituted of two main domains, a large N-terminal domain and a small C-terminal domain, separated by a cleft and linked by a hinge. <scene name='Sandbox2442/N-ter_domain/2'>The N-ter domain</scene> (residues 1-436) is made of a β-barrel and two β-sheets, flanked by α-helices, thus forming | ||
| + | <scene name='Sandbox2442/N-ter_domain/4'>an αβαβα five-layered structure</scene>. The <scene name='Sandbox2442/C-ter_domain/3'>C-ter domain</scene> (residues 440-550) forms a small | ||
| + | <scene name='Sandbox2442/C-ter_domain/4'>α+ β domain</scene>. | ||
| + | |||
| + | There are two conformational states, one when the enzyme is substrate-free, and the other one product-bound (whose structure is represented on this page). The transition is achieved by a 90° counterclockwise rotation of the <scene name='Sandbox2442/N-ter_c-ter/1'>C-domain relative to the N-domain</scene><ref name="ane">PMID:17513367</ref> | ||
| + | |||
| + | |||
| + | |||
| + | ===Sequence similarities=== | ||
| + | The firefly luciferase shares homologuous sequences and mechanisms with the peptide synthetases and acyl-CoA ligases, with a very few residues always conserved. | ||
| + | The firefly luciferase belongs to the super-family of acyl-adenylate-forming and thioester-forming enzymes. | ||
| + | |||
| + | But the firefly luciferase also have caracteristic sequences found in no other nucleotide-binding proteins, such as a motif responsible for the binding of ATP. | ||
| + | |||
| + | ===Active site=== | ||
| + | The firefly luciferase interacts with ATP, luciferin, an Acetyl-coenzyme A (CoA ) and O<sub>2</sub>. So differents groups of residues are part of the the ligand binding. | ||
| + | Those residues can be placed in the molecule by comparing the sequence of different enzymes of the same family. | ||
| + | The ATP binds to a <scene name='Sandbox2442/Atp_binding/3'>G-x-x-x-x-G-K-[STG]</scene> sequence which is a very disordered version of a classical mononucleotide-binding motif. <ref>PMID:8805533</ref> | ||
| + | The luciferin-binding site seems to be a depression between β sheet B and the barrel subdomain, but is not clearly visible on the structure. | ||
| + | The CoA-binding site does not have a classical motif of adenylate forming enzyme and this luciferase is the first member of a new superfamily to be studied. The adenylate forming reaction is likely to involve a few glycine or charged residues. | ||
| + | The others binding sites are not clearly defined yet but there are some conserved residues taking part in the binding or the catalysis: | ||
| + | |||
| + | * <scene name='Sandbox2442/Residue_thr_346/1'>Thr346</scene> which is the only residue with dihedral angles outside of the usual regions of the Ramachandran plot. This energetically unfavorable angles are usually in relation with residues having important functional roles. | ||
| + | |||
| + | * <scene name='Sandbox2442/Residue_asp_422/1'>Asp422</scene> is an invariant residue exposed to the solvant, and could make an hydrogen bond with the side chain of another well-conserved residue, the Tyr340. | ||
| + | |||
| + | * <scene name='Sandbox2442/Residue_ser_420/1'>Ser420</scene> has an hydroxyl group able to make hydrogen bond with the Asp422 and Gly421 which are close enough. | ||
| + | |||
| + | All the <scene name='Sandbox2442/Active_site/5'>conserved residues</scene> are either on the surface of the two domains on the side of the cleft or on the loop connecting the two domains. But this cleft is too big to accomodate on the substrates and is seems that the two domains only come closer around the products when they are formed. | ||
| + | |||
{{STRUCTURE_2d1s| PDB=2d1s ||SIZE=400| SCENE=Luciferase/2d1s/2 |CAPTION= 2d1s, resolution 1.30Å (<scene name='Luciferase/2d1s/2'>default scene</scene>). }} | {{STRUCTURE_2d1s| PDB=2d1s ||SIZE=400| SCENE=Luciferase/2d1s/2 |CAPTION= 2d1s, resolution 1.30Å (<scene name='Luciferase/2d1s/2'>default scene</scene>). }} | ||
| - | == | + | |
| + | === High-energy intermediate analogue=== | ||
Generally, firefly luciferases have some similarities with Acyl-CoA ligases and some peptide synthetases despite having different cellular effects. In fixing the structure of L. cruciata luciferase, the analog of a potent aminoacyl-tRNA synthetases (DLSA) was successfuly utilized to represent a stable oxyluciferin intermediate.<ref name="structure">PMID:16541080 </ref>.<br> | Generally, firefly luciferases have some similarities with Acyl-CoA ligases and some peptide synthetases despite having different cellular effects. In fixing the structure of L. cruciata luciferase, the analog of a potent aminoacyl-tRNA synthetases (DLSA) was successfuly utilized to represent a stable oxyluciferin intermediate.<ref name="structure">PMID:16541080 </ref>.<br> | ||
{{Link Toggle FancyCartoonHighQualityView}}.<br> | {{Link Toggle FancyCartoonHighQualityView}}.<br> | ||
The DLSA occupied the active site of the luciferase, which is composed of an α-helix (residues 248-260) and four short β-sheets (residues 286-289, 313-316, 339-342 and 351-353. Ile288 has been implicated as an important residue in determining the hydrophobicity of the active site environment, and through orientation of the product oxyluciferin, the bioluminescent colour. <ref name="structure" />. | The DLSA occupied the active site of the luciferase, which is composed of an α-helix (residues 248-260) and four short β-sheets (residues 286-289, 313-316, 339-342 and 351-353. Ile288 has been implicated as an important residue in determining the hydrophobicity of the active site environment, and through orientation of the product oxyluciferin, the bioluminescent colour. <ref name="structure" />. | ||
| - | + | {{STRUCTURE_2d1s| PDB=2d1s ||SIZE=400| SCENE=Luciferase/2d1s/2 |CAPTION= 2d1s, resolution 1.30Å (<scene name='Luciferase/2d1s/2'>default scene</scene>). }} | |
| + | |||
| + | ==Spectral difference with mutated luciferases== | ||
| + | |||
| + | The colour changes from the classical yellow-green colour to red with the substitution of a single amino acid, <scene name='Sandbox2442/Residue_ser_286/1'>Ser286</scene> to Asn, in the S286N mutant. The active site (and the residue Ile288) is less potent to effectue it's conformational change to the closed state, which was providing an extremely hydrophobic environment. This is then allowing an energy loss, and the product will emit lower energy light, with a wavelength moved to the red. | ||
| + | Further mutations proved that the <scene name='Sandbox2442/Ile_288/1'>amino acid residue at position 288</scene> is influencing the wavelength of the light emitted.<ref>PMID:16541080</ref> | ||
| + | |||
| + | |||
| + | {{STRUCTURE_2d1r| PDB=2d1r | SCENE= }} | ||
| + | |||
| + | ==Applications of the luciferase== | ||
| + | In genetic engineering, the Luciferase gene may be used as a good reporter gene, e.g. in expression vectors, for the sensibility it provides, its ease of use, instant quantification, "environment friendliness" and cost efficiency<ref>Giguère, V. (1991) Application of the firefly luciferase reporter gene. In: Methods in Molecular Biology, Vol. 7: Gene Transfer and Expression Protocols (E. J. Murray, ed.), The Humana Press Inc., Clifton, NJ, pp. 237-241. [http://www.springerprotocols.com/Abstract/doi/10.1385/0-89603-178-0:237]</ref>. | ||
| + | Luciferase can be used to measure ATP <ref>Hawronskyj J.-M, Measurement of ATP using firefly luminescence. European food and drink review. 1997, SUMMER, pp. 61-63 ISSN [http://cat.inist.fr/?aModele=afficheN&cpsidt=2717464 0955-4416]</ref> | ||
| + | It can also be used to study the action of general anesthetics<ref name="ane" />, which are inhibiting it. | ||
| + | The [http://en.wikipedia.org/wiki/Luciferase#Applications applications for luciferase] are very diverse. | ||
== Luciferase Control == | == Luciferase Control == | ||
As the structure of luciferases differ between species, so does the method of control over the bioluminescent reaction. In L. polyedrum, a marine dinoflagellate responsible for some red tides, a pH-dependant mechanism at the protein level appears to be responsible for control of bioluminescence. With fireflies however, the reaction is under at least some form of nervous control, with the insect controlling flashes through the use of nitric oxide <ref name="lights">PMID:11431567 </ref>. | As the structure of luciferases differ between species, so does the method of control over the bioluminescent reaction. In L. polyedrum, a marine dinoflagellate responsible for some red tides, a pH-dependant mechanism at the protein level appears to be responsible for control of bioluminescence. With fireflies however, the reaction is under at least some form of nervous control, with the insect controlling flashes through the use of nitric oxide <ref name="lights">PMID:11431567 </ref>. | ||
| - | == | + | ==Biology== |
| - | < | + | The [http://en.wikipedia.org/wiki/Bioluminescence bioluminescent] systems in the living organisms are very diverse (for example the luciferase has a different structure and catalyses a reaction in one step in the bacteria), so it is thought that they appeared separately in the course of evolution. |
| + | In general [http://en.wikipedia.org/wiki/Firefly fireflies] use bioluminescence to locate other individuals for mating, or to lure other species which are their preys. In the larvae, it is a warning signal for the predators, implying the presence of toxins. | ||
| + | Bioluminescence is thus utilized by several nocturnal japanese firely species during mate selection, with males and females illuminating equally. Several common signals appear to be used to communicate everything from "male awaiting a mate" to "female here". <ref name="main">PMID:8813052</ref> While the reaction is quite similiar to that of other bioluminescent luciferases, firefly luciferase has a unique structure in both the protein and luciferin required to produce the bioluminescence. In research, the firefly luciferase from ''Luciola cruciata'' is one of many commonly utilized for such purposes as such as sensing cellular ATP levels or visualizing the effects of a promoter sequence, among several others. | ||
| + | |||
==See Also== | ==See Also== | ||
| Line 27: | Line 98: | ||
*[http://www.ncbi.nlm.nih.gov/protein/CAA59282.1 NCBI protein entry on ''Photinus pyralis'' luciferase, the american firefly] | *[http://www.ncbi.nlm.nih.gov/protein/CAA59282.1 NCBI protein entry on ''Photinus pyralis'' luciferase, the american firefly] | ||
| + | |||
| + | ==References== | ||
| + | <references /> | ||
Revision as of 08:52, 26 November 2010
| |||||||||
| 2d1r, resolution 1.60Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Non-Standard Residues: | |||||||||
| Activity: | Photinus-luciferin 4-monooxygenase (ATP-hydrolyzing), with EC number 1.13.12.7 | ||||||||
| Related: | 2d1s, 2d1t | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Introduction
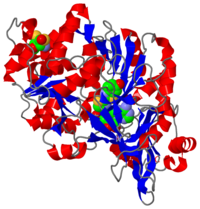
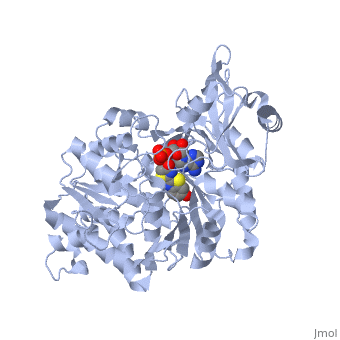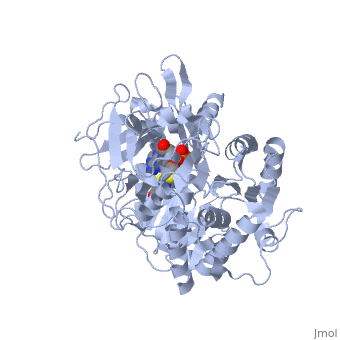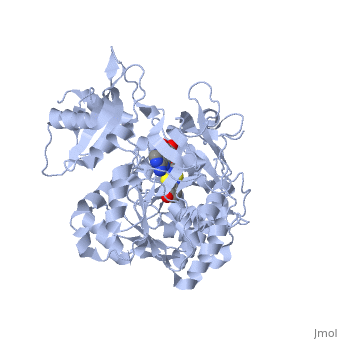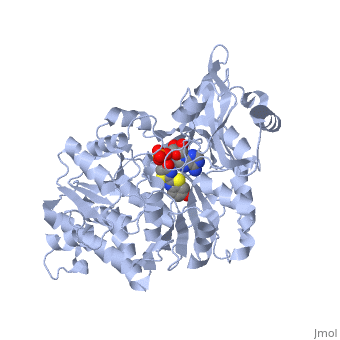
Luciferase is a class of enzymes producing light through the process of bioluminescence. This luciferase was extracted from Luciola cruciata. In the case of fireflies, it catalyses a reaction of adenylation and then an oxydative decarboxylation, changing luciferin to oxyluciferin and thus emitting light. This protein is constituted of two main domains separated by a cleft.
Chemical reaction
The enzyme catalyses the production of light, using two different conformation to catalyze two half-reactions (in the case of firefly Luciferase).
- luciferin + ATP → luciferyl adenylate + PPi
- luciferyl adenylate + O2 → + + light
The first half-reaction is an adenylation, the second one is an oxydative decarboxylation.
The production of light is achieved by the conversion of chemical energy into an emission of photons, resulting from the passage of the oxyluciferin's excitation state to a ground state.
The structure
2D1R is a 1 chain structure of sequence from Luciola cruciata. Full crystallographic information is available from OCA. The protein is shown here bound with two products, adenosine 5'-phosphate (AMP) and oxyluciferine.
It weights 62 kDa and is constituted of two main domains, a large N-terminal domain and a small C-terminal domain, separated by a cleft and linked by a hinge. (residues 1-436) is made of a β-barrel and two β-sheets, flanked by α-helices, thus forming
. The (residues 440-550) forms a small
.
There are two conformational states, one when the enzyme is substrate-free, and the other one product-bound (whose structure is represented on this page). The transition is achieved by a 90° counterclockwise rotation of the [1]
Sequence similarities
The firefly luciferase shares homologuous sequences and mechanisms with the peptide synthetases and acyl-CoA ligases, with a very few residues always conserved. The firefly luciferase belongs to the super-family of acyl-adenylate-forming and thioester-forming enzymes.
But the firefly luciferase also have caracteristic sequences found in no other nucleotide-binding proteins, such as a motif responsible for the binding of ATP.
Active site
The firefly luciferase interacts with ATP, luciferin, an Acetyl-coenzyme A (CoA ) and O2. So differents groups of residues are part of the the ligand binding. Those residues can be placed in the molecule by comparing the sequence of different enzymes of the same family. The ATP binds to a sequence which is a very disordered version of a classical mononucleotide-binding motif. [2] The luciferin-binding site seems to be a depression between β sheet B and the barrel subdomain, but is not clearly visible on the structure. The CoA-binding site does not have a classical motif of adenylate forming enzyme and this luciferase is the first member of a new superfamily to be studied. The adenylate forming reaction is likely to involve a few glycine or charged residues. The others binding sites are not clearly defined yet but there are some conserved residues taking part in the binding or the catalysis:
- which is the only residue with dihedral angles outside of the usual regions of the Ramachandran plot. This energetically unfavorable angles are usually in relation with residues having important functional roles.
- is an invariant residue exposed to the solvant, and could make an hydrogen bond with the side chain of another well-conserved residue, the Tyr340.
- has an hydroxyl group able to make hydrogen bond with the Asp422 and Gly421 which are close enough.
All the are either on the surface of the two domains on the side of the cleft or on the loop connecting the two domains. But this cleft is too big to accomodate on the substrates and is seems that the two domains only come closer around the products when they are formed.
| |||||||||
| 2d1s, resolution 1.30Å (). | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Non-Standard Residues: | |||||||||
| Activity: | Photinus-luciferin 4-monooxygenase (ATP-hydrolyzing), with EC number 1.13.12.7 | ||||||||
| Related: | 2d1r, 2d1t | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
High-energy intermediate analogue
Generally, firefly luciferases have some similarities with Acyl-CoA ligases and some peptide synthetases despite having different cellular effects. In fixing the structure of L. cruciata luciferase, the analog of a potent aminoacyl-tRNA synthetases (DLSA) was successfuly utilized to represent a stable oxyluciferin intermediate.[3].
.
The DLSA occupied the active site of the luciferase, which is composed of an α-helix (residues 248-260) and four short β-sheets (residues 286-289, 313-316, 339-342 and 351-353. Ile288 has been implicated as an important residue in determining the hydrophobicity of the active site environment, and through orientation of the product oxyluciferin, the bioluminescent colour. [3].
| |||||||||
| 2d1s, resolution 1.30Å (). | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Non-Standard Residues: | |||||||||
| Activity: | Photinus-luciferin 4-monooxygenase (ATP-hydrolyzing), with EC number 1.13.12.7 | ||||||||
| Related: | 2d1r, 2d1t | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Spectral difference with mutated luciferases
The colour changes from the classical yellow-green colour to red with the substitution of a single amino acid, to Asn, in the S286N mutant. The active site (and the residue Ile288) is less potent to effectue it's conformational change to the closed state, which was providing an extremely hydrophobic environment. This is then allowing an energy loss, and the product will emit lower energy light, with a wavelength moved to the red. Further mutations proved that the is influencing the wavelength of the light emitted.[4]
| |||||||||
| 2d1r, resolution 1.60Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Non-Standard Residues: | |||||||||
| Activity: | Photinus-luciferin 4-monooxygenase (ATP-hydrolyzing), with EC number 1.13.12.7 | ||||||||
| Related: | 2d1s, 2d1t | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Applications of the luciferase
In genetic engineering, the Luciferase gene may be used as a good reporter gene, e.g. in expression vectors, for the sensibility it provides, its ease of use, instant quantification, "environment friendliness" and cost efficiency[5]. Luciferase can be used to measure ATP [6] It can also be used to study the action of general anesthetics[1], which are inhibiting it. The applications for luciferase are very diverse.
Luciferase Control
As the structure of luciferases differ between species, so does the method of control over the bioluminescent reaction. In L. polyedrum, a marine dinoflagellate responsible for some red tides, a pH-dependant mechanism at the protein level appears to be responsible for control of bioluminescence. With fireflies however, the reaction is under at least some form of nervous control, with the insect controlling flashes through the use of nitric oxide [7].
Biology
The bioluminescent systems in the living organisms are very diverse (for example the luciferase has a different structure and catalyses a reaction in one step in the bacteria), so it is thought that they appeared separately in the course of evolution. In general fireflies use bioluminescence to locate other individuals for mating, or to lure other species which are their preys. In the larvae, it is a warning signal for the predators, implying the presence of toxins. Bioluminescence is thus utilized by several nocturnal japanese firely species during mate selection, with males and females illuminating equally. Several common signals appear to be used to communicate everything from "male awaiting a mate" to "female here". [8] While the reaction is quite similiar to that of other bioluminescent luciferases, firefly luciferase has a unique structure in both the protein and luciferin required to produce the bioluminescence. In research, the firefly luciferase from Luciola cruciata is one of many commonly utilized for such purposes as such as sensing cellular ATP levels or visualizing the effects of a promoter sequence, among several others.
See Also
- Japanese firefly Luciferase complexed with Oxyluciferin & AMP
- Luciferase
- Dinoflagellate luciferase
- Colored & Bioluminescent Proteins
- PyMOL
External Resources
References
- ↑ 1.0 1.1 Szarecka A, Xu Y, Tang P. Dynamics of firefly luciferase inhibition by general anesthetics: Gaussian and anisotropic network analyses. Biophys J. 2007 Sep 15;93(6):1895-905. Epub 2007 May 18. PMID:17513367 doi:10.1529/biophysj.106.102780
- ↑ Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996 Mar 15;4(3):287-98. PMID:8805533
- ↑ 3.0 3.1 Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Structural basis for the spectral difference in luciferase bioluminescence. Nature. 2006 Mar 16;440(7082):372-6. PMID:16541080 doi:10.1038/nature04542
- ↑ Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Structural basis for the spectral difference in luciferase bioluminescence. Nature. 2006 Mar 16;440(7082):372-6. PMID:16541080 doi:10.1038/nature04542
- ↑ Giguère, V. (1991) Application of the firefly luciferase reporter gene. In: Methods in Molecular Biology, Vol. 7: Gene Transfer and Expression Protocols (E. J. Murray, ed.), The Humana Press Inc., Clifton, NJ, pp. 237-241. [1]
- ↑ Hawronskyj J.-M, Measurement of ATP using firefly luminescence. European food and drink review. 1997, SUMMER, pp. 61-63 ISSN 0955-4416
- ↑ Trimmer BA, Aprille JR, Dudzinski DM, Lagace CJ, Lewis SM, Michel T, Qazi S, Zayas RM. Nitric oxide and the control of firefly flashing. Science. 2001 Jun 29;292(5526):2486-8. PMID:11431567 doi:10.1126/science.1059833
- ↑ Suzuki H, Sato Y, Fujiyama S, Ohba N. Biochemical systematics of Japanese fireflies of the subfamily Luciolinae and their flash communication systems. Biochem Genet. 1996 Jun;34(5-6):191-200. PMID:8813052
Proteopedia Page Contributors and Editors (what is this?)
Loïc Gazquez, Marion Bonazzi, Wayne Decatur, Alexander Berchansky, James Jones, David Canner, Michal Harel, Jaime Prilusky