Beta-glucosidase
From Proteopedia
| Line 62: | Line 62: | ||
==Additional Resources== | ==Additional Resources== | ||
For additional information, see: [[Carbohydrate Metabolism]] | For additional information, see: [[Carbohydrate Metabolism]] | ||
| + | |||
| + | ==3D structures of Beta-glucosidase== | ||
| + | |||
| + | [[3ahx]] – GBA – Clostridium cellulovorans<br /> | ||
| + | [[3ahy]] – GB – Trichoderma reesei<br /> | ||
| + | [[3abz]] – KmGB – Kluyveromyces marxianus<br /> | ||
| + | [[2x40]] – TnGB3B – Thermotoga neapolitana<br /> | ||
| + | [[3gno]] - JrGB residues 38-521 – Japanese rice<br /> | ||
| + | [[2rgl]], [[2rgm]] - JrGB residues 29-504<br /> | ||
| + | [[3f4v]], [[3f5j]], [[3f5k]], [[3f5l]] - JrGB7 (mutant) <br /> | ||
| + | [[3ahz]] – tGB – termite<br /> | ||
| + | [[3fiy]], [[3cmj]] - UbGB catalytic domain (mutant) – Uncultured bacteria<br /> | ||
| + | [[2o9p]], [[1bga]] – PpGBB – Paenibacillus polymyxa<br /> | ||
| + | [[2jfe]] - hGB cystolic – human<br /> | ||
| + | [[2e3z]] – PcGB – Phanerochaete crysosporium<br /> | ||
| + | [[2dga]] - wGB residues 1-520 – wheat<br /> | ||
| + | [[2vff]] – GB – Pyrococcus horikoshii<br /> | ||
| + | [[1oif]], [[1od0]] – TmGB catalytic domain - Thermotoga maritima<br /> | ||
| + | [[1ug6]] – GB - Thermus thermophilus<br /> | ||
| + | [[1hxj]], [[1e1e]] – ZmGB – Zea mays<br /> | ||
| + | [[1e4l]] - ZmGB (mutant) <br /> | ||
| + | [[1gon]] – SsGB – Streptomyces sp. <br /> | ||
| + | [[1qox]] – GB]] - Bacillus circulans<br /> | ||
| + | [[1tr1]] - BpGBB (mutant)]] - Bacillus polymyxa<br /> | ||
| + | [[1cbg]] – GB cyanogenic – White clover<br /> | ||
| + | [[3aiu]] - rGB residues 50-568 – rye | ||
| + | |||
| + | ===Beta-glucosidase complex with sugar=== | ||
| + | |||
| + | [[3ai0]] – tGB + glucoside<br /> | ||
| + | [[3ac0]] - KmGB + glucoside<br /> | ||
| + | [[2x41]], [[2x42]] - TnGB + glucoside<br /> | ||
| + | [[3air]] – wGB residues 50-569 + glucoside + dinitrophenol<br /> | ||
| + | [[3ais]] - wGB residues 50-569 (mutant) + glucoside + aglycone<br /> | ||
| + | [[3aiq]] - wGB residues 50-569 + aglycone<br /> | ||
| + | [[3aiv]] - rGB residues 50-568 + aglycone<br /> | ||
| + | [[3aiw]] - rGB residues 50-568 + glucoside + dinitrophenol<br /> | ||
| + | [[3gnp]], [[3gnr]] - JrGB residues 38-521 + glucoside<br /> | ||
| + | [[3aht]], [[3ahv]] – JrGB7 (mutant) + saccharide<br /> | ||
| + | [[1oin]] - TmGBA + glucoside<br /> | ||
| + | [[3fiz]], [[3fj0]] - UbGB residues 18-482 (mutant) + glucoside<br /> | ||
| + | [[2zox]] - hGB cystolic + glucoside<br /> | ||
| + | [[2o9s]], [[2o9r]], [[2z1s]] – PpGBB + saccharide<br /> | ||
| + | [[2o9t]] - PpGBB + glucoside<br /> | ||
| + | [[1uyq]] - PpGBB (mutant) + glucoside<br /> | ||
| + | [[1bgg]] - PpGBB + gluconate<br /> | ||
| + | [[2jie]] - BpGBB + glucoside<br /> | ||
| + | [[1e4i]] - BpGBB (mutant) + glucoside<br /> | ||
| + | [[2e40]] – PcGB + gluconolactone<br /> | ||
| + | [[1v08]] – ZmGB + gluco-tetrazole<br /> | ||
| + | [[1e1f]] - ZmGB + glucoside<br /> | ||
| + | [[1h49]], [[1e4n]], [[1e56]] - ZmGB (mutant) + aglycone<br /> | ||
| + | [[1gnx]] - SsGB + saccharide - Sulfolobus solfataricus | ||
| + | |||
| + | ===Beta-glucosidase complex with inhibitor=== | ||
| + | |||
| + | [[2wbg]], [[2wc3]], [[2wc4]], [[2vrj]], [[2jal]], [[2j75]], [[2j77]], [[2j78]], [[2j79]], [[2j7b]], [[2j7d]], [[2j7e]], [[2j7f]], [[2j7g]], [[2j7h]], [[2j7c]], [[2ces]], [[2cet]], [[2cbu]], [[2cbv]], [[1uz1]], [[1wj3]], [[1oim]] – TmGBA + inhibitor<br /> | ||
| + | [[2cer]] – SsGB + inhibitor <br /> | ||
| + | [[1e55]] - ZmGB (mutant) + inhibitor | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 09:51, 15 May 2011
A β-glucosidase is an enzyme which catalyses the hydrolysis of terminal non-reducing residues in β-glucosides (EC number : 3.2.1.21). In the case of 2VRJ, it comes from Thermotoga maritima which is a rod-shaped bacterium belonging to the order of Thermotogates. This bacterium was originally isolated from geothermal heated marine sediments. 2VRJ is here is in complex with an inhibitor called N-octyl-5-deoxy66-oxa-N-carbamoylcalystegine [1].
Contents |
General action as biocatalyst
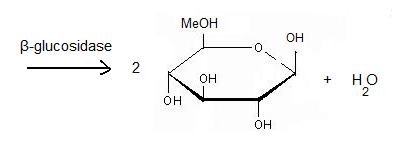
It acts on the β(1-4) bond linking two glucose residues or glucose-substituted molecules. The action of the enzyme on such glucosides results in the release of units of glucose. For instance, hydrolysis of cellobiose catalysed by a β-glucosidase releases two glucoses [2].
β-glucosidases can also be called β-D-glucoside glucohydrolases or cellobiases.
2VRJ
Structure and function
| |||||||||||
Hydrolysis of terminal non-reducing residues in β-glucosides
There are two ways to hydrolyse the terminal non-reducing residues in β-glucosides which implicate the two glutamate residues and a molecule of water [7]. Water which is an amphoter, is here used as a base for the nucleophilic attack on the positively charged anomeric carbon.
The general equation of the chemical reaction is :
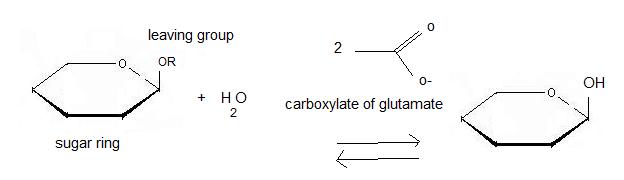
Inverting glycoside hydrolases
Inverting glycoside hydrolases lead to an inversion of the anomeric configuration to create an alpha configuration. The steps of the reaction are like the mechanism of nucleophilic substitution S2N. It is an one step process: the nucleophile (water) the anomeric carbon with simultaneous expulsion of the leaving group (OR). Bond making takes place at the same time as bond breaking. Such a mechanism is called concerted reaction. The distance between the two carboxylates is about 10.5 angströms.
Retaining glycoside hydrolases
Retaining glycoside hydrolyses occur in two steps: the first step, called glycosylation leads to the release of the leaving group and the creation of a carbocation. Subsequently, water attacks this last one. The second step, called deglycosylation consists of OR- nucleophilic attack on the intermediate and allows the deglycosylation of the enzyme. In this case, there are two transition states involved. The distance between the two carboxylates for this mechanism is about 5.5 angströms. For 2VRJ the distance between its two glutamates is about angströms. So we can say that 2VRJ seems to be a retaining enzyme.
NB: The values of the pH and the nature of the solvent play a main role in the rate of the reaction.
Glutamates are directly involved in the catalytic reaction but asparagine is used to stabilise the structure.
Other use of β-glucosidases
β-glucosidase is now used for the synthesis of biofuel. Wood is an abundant and renewable energy which can be changed into bioethanol thanks to enzymatic hydrolysis. This synthesis needs five steps. First it is pre-hydrolysis. The structure is divided into lignin and (hemi)cellulose. The cellulase can better access the structure to act on it. The second step: hydrolysis is the most important. A cellulase is a complex of 3 enzymes which act together to hydrolyse cellulose: Endoglucanase breaks the chain in the middle of the molecular structure of cellulose. Exoglucanase binds an available end of the chain and isolates it. Then units of cellobiose are cut (two units of glucose which are together). To finish, β-glucosidase divides cellobiose into two glucoses. When they ferment, they become ethanol. The final product is obtained thanks to fermentation, distillation and deshydratation.
Additional Resources
For additional information, see: Carbohydrate Metabolism
3D structures of Beta-glucosidase
3ahx – GBA – Clostridium cellulovorans
3ahy – GB – Trichoderma reesei
3abz – KmGB – Kluyveromyces marxianus
2x40 – TnGB3B – Thermotoga neapolitana
3gno - JrGB residues 38-521 – Japanese rice
2rgl, 2rgm - JrGB residues 29-504
3f4v, 3f5j, 3f5k, 3f5l - JrGB7 (mutant)
3ahz – tGB – termite
3fiy, 3cmj - UbGB catalytic domain (mutant) – Uncultured bacteria
2o9p, 1bga – PpGBB – Paenibacillus polymyxa
2jfe - hGB cystolic – human
2e3z – PcGB – Phanerochaete crysosporium
2dga - wGB residues 1-520 – wheat
2vff – GB – Pyrococcus horikoshii
1oif, 1od0 – TmGB catalytic domain - Thermotoga maritima
1ug6 – GB - Thermus thermophilus
1hxj, 1e1e – ZmGB – Zea mays
1e4l - ZmGB (mutant)
1gon – SsGB – Streptomyces sp.
1qox – GB]] - Bacillus circulans
1tr1 - BpGBB (mutant)]] - Bacillus polymyxa
1cbg – GB cyanogenic – White clover
3aiu - rGB residues 50-568 – rye
Beta-glucosidase complex with sugar
3ai0 – tGB + glucoside
3ac0 - KmGB + glucoside
2x41, 2x42 - TnGB + glucoside
3air – wGB residues 50-569 + glucoside + dinitrophenol
3ais - wGB residues 50-569 (mutant) + glucoside + aglycone
3aiq - wGB residues 50-569 + aglycone
3aiv - rGB residues 50-568 + aglycone
3aiw - rGB residues 50-568 + glucoside + dinitrophenol
3gnp, 3gnr - JrGB residues 38-521 + glucoside
3aht, 3ahv – JrGB7 (mutant) + saccharide
1oin - TmGBA + glucoside
3fiz, 3fj0 - UbGB residues 18-482 (mutant) + glucoside
2zox - hGB cystolic + glucoside
2o9s, 2o9r, 2z1s – PpGBB + saccharide
2o9t - PpGBB + glucoside
1uyq - PpGBB (mutant) + glucoside
1bgg - PpGBB + gluconate
2jie - BpGBB + glucoside
1e4i - BpGBB (mutant) + glucoside
2e40 – PcGB + gluconolactone
1v08 – ZmGB + gluco-tetrazole
1e1f - ZmGB + glucoside
1h49, 1e4n, 1e56 - ZmGB (mutant) + aglycone
1gnx - SsGB + saccharide - Sulfolobus solfataricus
Beta-glucosidase complex with inhibitor
2wbg, 2wc3, 2wc4, 2vrj, 2jal, 2j75, 2j77, 2j78, 2j79, 2j7b, 2j7d, 2j7e, 2j7f, 2j7g, 2j7h, 2j7c, 2ces, 2cet, 2cbu, 2cbv, 1uz1, 1wj3, 1oim – TmGBA + inhibitor
2cer – SsGB + inhibitor
1e55 - ZmGB (mutant) + inhibitor
References
- ↑ Aguilar M, Gloster TM, Garcia-Moreno MI, Ortiz Mellet C, Davies GJ, Llebaria A, Casas J, Egido-Gabas M, Garcia Fernandez JM. Molecular basis for beta-glucosidase inhibition by ring-modified calystegine analogues. Chembiochem. 2008 Nov 3;9(16):2612-8. PMID:18833549 doi:10.1002/cbic.200800451
- ↑ http://en.wikipedia.org/wiki/B-glucosidase
- ↑ Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995 Sep 15;3(9):853-9. PMID:8535779
- ↑ http://www.ebi.ac.uk/interpro/IEntry?ac=IPR018120#PUB00002205
- ↑ http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/CSA/CSA_Site_Wrapper.pl?pdb=2vrj
- ↑ Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995 Sep 15;3(9):853-9. PMID:8535779
- ↑ http://www.cazy.org/fam/ghf_INV_RET.html#3
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Muriel Breteau, Alexander Berchansky, Joel L. Sussman, David Canner