Aminopeptidase
From Proteopedia
| Line 2: | Line 2: | ||
{{STRUCTURE_1xjo| PDB=1xjo | SIZE=300| SCENE= |right|CAPTION=Aminopeptidase [[1xjo]] }} | {{STRUCTURE_1xjo| PDB=1xjo | SIZE=300| SCENE= |right|CAPTION=Aminopeptidase [[1xjo]] }} | ||
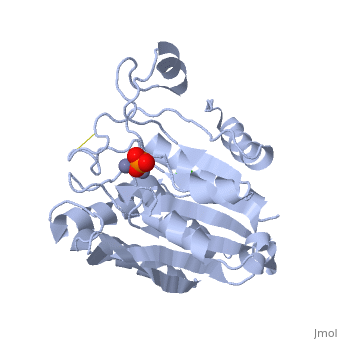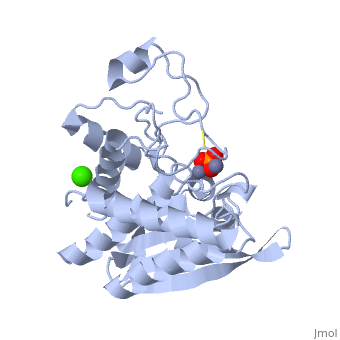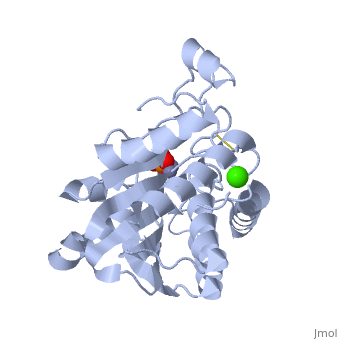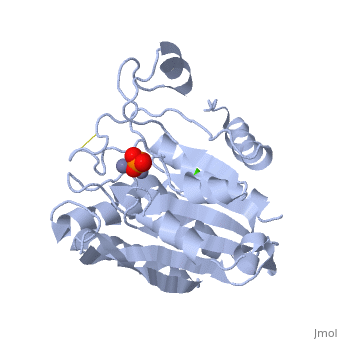
| - | '''Aminopeptidases''' catalyze a release of an N-terminal amino acid from a peptide, amide, or arylamide. | + | [[Aminopeptidase|Aminopeptidases]] (AP) are metal - mostly Zn-dependent enzymes involved in the digestion of proteins. Cytosol AP (Cyt-AP), deblocking AP (DAP) and AP N (APN) remove N-terminal amino acids. The AP are classified by the amino acid which they hydrolyze. Other types of AP are: Cold-activated AP (Col-AP), Heat stable AP from Thermus thermophilus (AmpT), AP from Staphylococcus aureus (AmpS) and SGAP from Stereomyces griseus. '''Aminopeptidases''' catalyze a release of an N-terminal amino acid from a peptide, amide, or arylamide. The images at the left and at the right correspond to one representative Aminopeptidase, ''i.e.'' the crystal structure of ''Stereomyces griesus'' aminopeptidase ([[1xjo]]). |
| + | |||
| + | {{TOC limit|limit=2}} | ||
| + | |||
| + | == 3D Structures of Aminopeptidase == | ||
| + | |||
| + | ===Cysteine aminopeptidase=== | ||
| + | |||
| + | [[3pw3]] – Cys-AP – ''Parabacteroides distasonis'' | ||
| + | |||
| + | ===Glutamic acid aminopeptidase=== | ||
| + | |||
| + | [[2wyr]] – PhGlu-AP+Co – ''Pyrococcus horikoshii''<br /> | ||
| + | [[3kl9]] – Glu-AP – ''Streptococcus pneumoniae'' | ||
| + | |||
| + | ===Alanine aminopeptidase=== | ||
| + | |||
| + | [[3ebg]] – PfAla-AP - ''Plasmodium falciparum''<br /> | ||
| + | [[3ebh]] - PfAla-AP+bestatin<br /> | ||
| + | [[3ebi]] - PfAla-AP+dipeptide analog | ||
| + | |||
| + | ===Proline aminopeptidase=== | ||
| + | |||
| + | [[3ovk]] – Xaa Pro-AP – ''Streptococcus pyogenes''<br /> | ||
| + | [[3il0]] - Xaa Pro-AP – ''Streptococcus thermophilus''<br /> | ||
| + | [[2v3x]], [[2v3y]], [[2v3z]] - Xaa EcPro-AP (mutant)+tripeptide<br /> | ||
| + | [[1w7v]], [[2bh3]] - Xaa EcPro-AP+Zn+Mg+polypeptide<br /> | ||
| + | [[2bn7]] - Xaa EcPro-AP+Zn+Mg+Mn+polypeptide<br /> | ||
| + | [[2bha]], [[2bhd]] - Xaa EcPro-AP+Mg+polypeptide<br /> | ||
| + | [[1a16]] - Xaa EcPro-AP+Mn+polypeptide<br /> | ||
| + | [[1wbq]], [[2bhb]] - Xaa EcPro-AP+Zn+Mg<br /> | ||
| + | [[2bhc]] - Xaa EcPro-AP+Na+Mg<br /> | ||
| + | [[1wl6]] - Xaa EcPro-AP+Mg<br /> | ||
| + | [[1wi9]], [[1m35]], [[1jaw]] - Xaa EcPro-AP+Mn<br /> | ||
| + | [[1wlr]] - Xaa EcPro-AP<br /> | ||
| + | [[2bws]], [[2bwt]], [[2bwu]], [[2bwv]], [[2bww]], [[2bwx]], [[2bwy]] - Xaa EcPro-AP (mutant)<br /> | ||
| + | [[1w2m]] - Xaa EcPro-AP+Ca<br /> | ||
| + | [[1n51]] – Xaa EcPro-AP+apstatin<br /> | ||
| + | [[2zsg]] – X TmPro-AP – ''Thermatoga maritime''<br /> | ||
| + | [[3ctz]] – X hPro-AP – human<br /> | ||
| + | [[1x2b]], [[1x2e]], [[1wm1]] – SmPro-AP + inhibitor – ''Serratia marcescens''<br /> | ||
| + | [[1qtr]] – SmPro-AP<br /> | ||
| + | [[1xqv]] – TaPro-AP (mutant) – ''Thermoplasma acidophilum''<br /> | ||
| + | [[1xqw]], [[1xqx]], [[1xqy]], [[1xrl]], [[1xrm]], [[1xrn]], [[1xro]], [[1xrp]], [[1xrq]], [[1xrr]] – TaPro-AP+polypeptide<br /> | ||
| + | |||
| + | ===Leucine aminopeptidase=== | ||
| + | |||
| + | [[3jru]] – Leu-AP – ''Xanthomonas oryzae''<br /> | ||
| + | [[2hc9]], [[2hb6]] – Leu-AP – ''Caenorhabditis elegans''<br /> | ||
| + | [[2ewb]] – bLeu-AP+zofenoprilat – bovine<br /> | ||
| + | [[1lam]], [[1lap]] – bLue-AP<br /> | ||
| + | [[1bpm]], [[1bpn]] – bLue-AP+Zn+Mg<br /> | ||
| + | [[1lan]], [[1lcp]] – bLue-AP+leucine derivative<br /> | ||
| + | [[1bll]] – bLue-AP+amastatin<br /> | ||
| + | [[2xdt]] – hLeu-AP soluble domain <br /> | ||
| + | [[3h8e]], [[3h8f]], [[3h8g]] – Leu-AP – ''Pseudomonas putida''<br /> | ||
| + | [[3kqx]], [[3kqz]], [[3kr4]], [[3kr5]] – PfLeu-AP <br /> | ||
| + | [[3fh4]], [[1rtq]], [[2dea]] – VpLeu-AP – Vibrio proteolyticus<br /> | ||
| + | [[3b35]], [[3b3t]], [[3b3v]], [[2anp]] - VpLeu-AP (mutant)<br /> | ||
| + | [[3b3c]], [[3b3s]], [[3b3w]] - VpLeu-AP (mutant)+Leu derivative<br /> | ||
| + | [[1ft7]] - VpLeu-AP +Leu derivative<br /> | ||
| + | [[2nyq]], [[2iq6]] – VpLeu-AP+polypeptide<br /> | ||
| + | [[1lok]] – VpLeu-AP+Tris<br /> | ||
| + | [[2prq]] – VpLeu-AP+Co<br /> | ||
| + | [[1xry]], [[1txr]] – VpLeu-AP+bestatin<br /> | ||
| + | [[1gyt]] – EcLeu-AP | ||
| + | |||
| + | ===Methionine aminopeptidase=== | ||
| + | |||
| + | [[3mr1]], [[3mx6]] – Met-AP+Mn – ''Rickettsia prowazekii''<br /> | ||
| + | [[2dfi]], [[1xgm]], [[1xgn]], [[1xgo]], [[1xgs]] – PfMet-AP+Co – ''Pyrococcus furiosus''<br /> | ||
| + | [[1wkm]] - PfMet-AP+Mn<br /> | ||
| + | [[3iu7]] – MtMet-AP+Mn +A02 – ''Mycobacterium tuberculosis''<br /> | ||
| + | [[3iu8]], [[3iu9]] - MtMet-AP+Ni + inhibitor<br /> | ||
| + | [[1yj3]] - MtMet-AP+Co<br /> | ||
| + | [[1y1n]] - MtMet-AP+K<br /> | ||
| + | [[3fm3]] – EncMet-AP – ''Encephalitozoon cuniculi''<br /> | ||
| + | [[3fmq]], [[3fmr]] - EncMet-AP+angiogenesis inhibitor<br /> | ||
| + | [[3d27]], [[2q92]], [[2q93]], [[2q94]], [[2q95]], [[2q96]], [[2p98]], [[2p99]], [[2p9a]], [[2gu4]], [[2gu5]], [[2gu6]], [[2evc]], [[2evm]], [[2evo]], [[2bbv]], [[1xnz]] – EcMet-AP+Mn+inhibitor<br /> | ||
| + | [[2gg0]], [[2gg2]], [[2gg3]], [[2gg5]], [[2gg7]], [[2gg8]], [[2gg9]], [[2ggb]], [[2ggc]] - EcMet-AP+Co+inhibitor<br /> | ||
| + | [[1c21]], [[1c22]], [[1c23]], [[1c24]] - EcMet-AP +Co + methionine derivative<br /> | ||
| + | [[1c27]] - EcMet-AP +Co +norleucine<br /> | ||
| + | [[2ea2]], [[2ea4]], [[2ga2]], [[2nq6]], [[2nq7]], [[1yw7]], [[1yw8]], [[1yw9]] - hMet-AP+Mn+inhibitor<br /> | ||
| + | [[2gtx]], [[2gu7]] – EcMet-AP<br /> | ||
| + | [[1yvm]] – EcMet-AP (mutant)+Co+thiabendazole<br /> | ||
| + | [[1mat]] – EcMet-AP+Co<br /> | ||
| + | [[2mat]], [[4mat]] - EcMet-AP (mutant)+Co<br /> | ||
| + | [[3mat]] - EcMet-AP (mutant)+Co+bestatin derivative<br /> | ||
| + | [[2b3h]], [[2b3k]] - hMet-AP+Co<br /> | ||
| + | [[1boa]] - hMet-AP+Co+ angiogenesis inhibitor<br /> | ||
| + | [[2b3l]] – hMet-AP<br /> | ||
| + | [[1kq0]], [[1kq9]] – hMet-AP+methionine<br /> | ||
| + | [[2gz5]], [[2adu]], [[1qzy]], [[1b59]], [[1b6a]], [[1bn5]] – hMet-AP+Co+inhibitor<br /> | ||
| + | [[1r58]], [[1r5g]], [[1r5h]] - hMet-AP+Mn+inhibitor<br /> | ||
| + | [[2g6p]] - hMet-AP truncated+Mn+inhibitor<br /> | ||
| + | [[1qxw]], [[1qxy]], [[1qxz]] – Met-AP+Co+inhibitor - ''Staphylococcus aureus''<br /> | ||
| + | [[1o0x]] – TmMet-AP | ||
| + | |||
| + | ===Aspartic acid aminopeptidase=== | ||
| + | |||
| + | [[3l6s]] – hAsp-AP+aspartic hydroxamate | ||
| + | |||
| + | ===Asparagine aminopeptidase=== | ||
| + | |||
| + | [[3c17]], [[2zak]] – EcAsn-AP (mutant)<br /> | ||
| + | [[2zal]] – EcAsn-AP+Asp<br /> | ||
| + | [[2gez]] – Asn-AP – ''Lupinus luteus'' | ||
| + | |||
| + | ===Serine aminopeptidase=== | ||
| + | |||
| + | [[1b65]] – Ser-AP – ''Ochrobactrum anthropi'' | ||
| + | |||
| + | ===Cytosolic aminopeptidase=== | ||
| + | |||
| + | [[3pei]] – Cyt-AP – ''Francisella tularensis''<br /> | ||
| + | [[3kzw]] – Cyt-AP – ''Staphylococcus aureus''<br /> | ||
| + | [[3ij3]] – Cyt-AP – ''Coxiella burnetii'' | ||
| + | |||
| + | ===Aminopeptidase N=== | ||
| + | |||
| + | [[3ked]] – EcAPN+2,4-diaminobutyric acid – ''Escherichia coli''<br /> | ||
| + | [[2hpo]], [[2dq6]] – EcAPN<br /> | ||
| + | [[2hpt]], [[2dqm]] – EcAPN+bestatin<br /> | ||
| + | [[2zxg]] – EcAPN+transition state analog<br /> | ||
| + | [[3b2p]], [[3b2x]], [[3b34]], [[3b37]], [[3b3b]] – EcAPN+amino acid<br /> | ||
| + | [[2gtq]] – APN – ''Neisseria meningitides'' | ||
| + | |||
| + | ===Non-specific aminopeptidase=== | ||
| + | |||
| + | [[2ek8]] – AnAP – Aneurinibacillus<br /> | ||
| + | [[2ek9]] – AnAP+bestatin<br /> | ||
| + | [[1y0r]], [[1xfo]] – PhAP<br /> | ||
| + | [[1y0y]] – PhAP+amastatin<br /> | ||
| + | [[1amp]] - VpAP<br /> | ||
| + | [[1cp6]], [[1igb]] – VpAP+inhibitor | ||
| + | |||
| + | ===Cold-activated aminopeptidase=== | ||
| + | |||
| + | [[3cia]] – Col-AP – ''Colwellia psychrerythraea'' | ||
| + | |||
| + | ===Deblocking aminopeptidase=== | ||
| + | |||
| + | [[2gre]] – DAP – ''Bacillus cereus'' | ||
| + | |||
| + | ===Heat stable aminopeptidase=== | ||
| + | |||
| + | [[2ayi]] – AmpT – ''Thermus thermophilus'' | ||
| + | |||
| + | ===Aminopeptidase from ''Staphylococcus aureus''=== | ||
| + | |||
| + | [[1zjc]] - AmpS | ||
| + | |||
| + | ===''Stereomyces griesus'' aminopeptidase=== | ||
| + | |||
| + | [[1xjo]], [[1cp7]] - SGAP | ||
| + | [[1qq9]], [[1f2o]], [[1f2p]], [[1tf8]], [[1tf9]], [[1tkf]], [[1tkh]], [[1tkj]], [[1xbu]] - SGAP + amino acid | ||
| + | |||
{{Clear}} | {{Clear}} | ||
== ''S. griseus'' aminopeptidase == | == ''S. griseus'' aminopeptidase == | ||
Revision as of 12:09, 30 December 2010
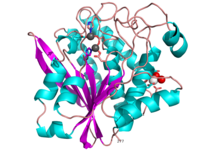
Aminopeptidases (AP) are metal - mostly Zn-dependent enzymes involved in the digestion of proteins. Cytosol AP (Cyt-AP), deblocking AP (DAP) and AP N (APN) remove N-terminal amino acids. The AP are classified by the amino acid which they hydrolyze. Other types of AP are: Cold-activated AP (Col-AP), Heat stable AP from Thermus thermophilus (AmpT), AP from Staphylococcus aureus (AmpS) and SGAP from Stereomyces griseus. Aminopeptidases catalyze a release of an N-terminal amino acid from a peptide, amide, or arylamide. The images at the left and at the right correspond to one representative Aminopeptidase, i.e. the crystal structure of Stereomyces griesus aminopeptidase (1xjo).
3D Structures of Aminopeptidase
Cysteine aminopeptidase
3pw3 – Cys-AP – Parabacteroides distasonis
Glutamic acid aminopeptidase
2wyr – PhGlu-AP+Co – Pyrococcus horikoshii
3kl9 – Glu-AP – Streptococcus pneumoniae
Alanine aminopeptidase
3ebg – PfAla-AP - Plasmodium falciparum
3ebh - PfAla-AP+bestatin
3ebi - PfAla-AP+dipeptide analog
Proline aminopeptidase
3ovk – Xaa Pro-AP – Streptococcus pyogenes
3il0 - Xaa Pro-AP – Streptococcus thermophilus
2v3x, 2v3y, 2v3z - Xaa EcPro-AP (mutant)+tripeptide
1w7v, 2bh3 - Xaa EcPro-AP+Zn+Mg+polypeptide
2bn7 - Xaa EcPro-AP+Zn+Mg+Mn+polypeptide
2bha, 2bhd - Xaa EcPro-AP+Mg+polypeptide
1a16 - Xaa EcPro-AP+Mn+polypeptide
1wbq, 2bhb - Xaa EcPro-AP+Zn+Mg
2bhc - Xaa EcPro-AP+Na+Mg
1wl6 - Xaa EcPro-AP+Mg
1wi9, 1m35, 1jaw - Xaa EcPro-AP+Mn
1wlr - Xaa EcPro-AP
2bws, 2bwt, 2bwu, 2bwv, 2bww, 2bwx, 2bwy - Xaa EcPro-AP (mutant)
1w2m - Xaa EcPro-AP+Ca
1n51 – Xaa EcPro-AP+apstatin
2zsg – X TmPro-AP – Thermatoga maritime
3ctz – X hPro-AP – human
1x2b, 1x2e, 1wm1 – SmPro-AP + inhibitor – Serratia marcescens
1qtr – SmPro-AP
1xqv – TaPro-AP (mutant) – Thermoplasma acidophilum
1xqw, 1xqx, 1xqy, 1xrl, 1xrm, 1xrn, 1xro, 1xrp, 1xrq, 1xrr – TaPro-AP+polypeptide
Leucine aminopeptidase
3jru – Leu-AP – Xanthomonas oryzae
2hc9, 2hb6 – Leu-AP – Caenorhabditis elegans
2ewb – bLeu-AP+zofenoprilat – bovine
1lam, 1lap – bLue-AP
1bpm, 1bpn – bLue-AP+Zn+Mg
1lan, 1lcp – bLue-AP+leucine derivative
1bll – bLue-AP+amastatin
2xdt – hLeu-AP soluble domain
3h8e, 3h8f, 3h8g – Leu-AP – Pseudomonas putida
3kqx, 3kqz, 3kr4, 3kr5 – PfLeu-AP
3fh4, 1rtq, 2dea – VpLeu-AP – Vibrio proteolyticus
3b35, 3b3t, 3b3v, 2anp - VpLeu-AP (mutant)
3b3c, 3b3s, 3b3w - VpLeu-AP (mutant)+Leu derivative
1ft7 - VpLeu-AP +Leu derivative
2nyq, 2iq6 – VpLeu-AP+polypeptide
1lok – VpLeu-AP+Tris
2prq – VpLeu-AP+Co
1xry, 1txr – VpLeu-AP+bestatin
1gyt – EcLeu-AP
Methionine aminopeptidase
3mr1, 3mx6 – Met-AP+Mn – Rickettsia prowazekii
2dfi, 1xgm, 1xgn, 1xgo, 1xgs – PfMet-AP+Co – Pyrococcus furiosus
1wkm - PfMet-AP+Mn
3iu7 – MtMet-AP+Mn +A02 – Mycobacterium tuberculosis
3iu8, 3iu9 - MtMet-AP+Ni + inhibitor
1yj3 - MtMet-AP+Co
1y1n - MtMet-AP+K
3fm3 – EncMet-AP – Encephalitozoon cuniculi
3fmq, 3fmr - EncMet-AP+angiogenesis inhibitor
3d27, 2q92, 2q93, 2q94, 2q95, 2q96, 2p98, 2p99, 2p9a, 2gu4, 2gu5, 2gu6, 2evc, 2evm, 2evo, 2bbv, 1xnz – EcMet-AP+Mn+inhibitor
2gg0, 2gg2, 2gg3, 2gg5, 2gg7, 2gg8, 2gg9, 2ggb, 2ggc - EcMet-AP+Co+inhibitor
1c21, 1c22, 1c23, 1c24 - EcMet-AP +Co + methionine derivative
1c27 - EcMet-AP +Co +norleucine
2ea2, 2ea4, 2ga2, 2nq6, 2nq7, 1yw7, 1yw8, 1yw9 - hMet-AP+Mn+inhibitor
2gtx, 2gu7 – EcMet-AP
1yvm – EcMet-AP (mutant)+Co+thiabendazole
1mat – EcMet-AP+Co
2mat, 4mat - EcMet-AP (mutant)+Co
3mat - EcMet-AP (mutant)+Co+bestatin derivative
2b3h, 2b3k - hMet-AP+Co
1boa - hMet-AP+Co+ angiogenesis inhibitor
2b3l – hMet-AP
1kq0, 1kq9 – hMet-AP+methionine
2gz5, 2adu, 1qzy, 1b59, 1b6a, 1bn5 – hMet-AP+Co+inhibitor
1r58, 1r5g, 1r5h - hMet-AP+Mn+inhibitor
2g6p - hMet-AP truncated+Mn+inhibitor
1qxw, 1qxy, 1qxz – Met-AP+Co+inhibitor - Staphylococcus aureus
1o0x – TmMet-AP
Aspartic acid aminopeptidase
3l6s – hAsp-AP+aspartic hydroxamate
Asparagine aminopeptidase
3c17, 2zak – EcAsn-AP (mutant)
2zal – EcAsn-AP+Asp
2gez – Asn-AP – Lupinus luteus
Serine aminopeptidase
1b65 – Ser-AP – Ochrobactrum anthropi
Cytosolic aminopeptidase
3pei – Cyt-AP – Francisella tularensis
3kzw – Cyt-AP – Staphylococcus aureus
3ij3 – Cyt-AP – Coxiella burnetii
Aminopeptidase N
3ked – EcAPN+2,4-diaminobutyric acid – Escherichia coli
2hpo, 2dq6 – EcAPN
2hpt, 2dqm – EcAPN+bestatin
2zxg – EcAPN+transition state analog
3b2p, 3b2x, 3b34, 3b37, 3b3b – EcAPN+amino acid
2gtq – APN – Neisseria meningitides
Non-specific aminopeptidase
2ek8 – AnAP – Aneurinibacillus
2ek9 – AnAP+bestatin
1y0r, 1xfo – PhAP
1y0y – PhAP+amastatin
1amp - VpAP
1cp6, 1igb – VpAP+inhibitor
Cold-activated aminopeptidase
3cia – Col-AP – Colwellia psychrerythraea
Deblocking aminopeptidase
2gre – DAP – Bacillus cereus
Heat stable aminopeptidase
2ayi – AmpT – Thermus thermophilus
Aminopeptidase from Staphylococcus aureus
1zjc - AmpS
Stereomyces griesus aminopeptidase
1xjo, 1cp7 - SGAP 1qq9, 1f2o, 1f2p, 1tf8, 1tf9, 1tkf, 1tkh, 1tkj, 1xbu - SGAP + amino acid
S. griseus aminopeptidase
|
S. griseus aminopeptidase (SGAP) cleaves the N-terminal amino acid from a peptide or protein, and is specific for larger hydrophobic acids, especially leucine. No cleavage occurs if the next residue is proline.
The of the enzyme contains two Zn2+ ions with His85 and Asp160 as ligands for one ion, and Glu132 and His247 as ligands for the second ion. Asp97 is a common ligand to both ions. What appears to be a phosphate anion is bound to both zinc atoms, replacing the water molecule/hydroxide ion normally found in this class of enzyme.
Additional Resources
For additional information, see: Amino Acid Synthesis & Metabolism
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Joel L. Sussman, Eran Hodis