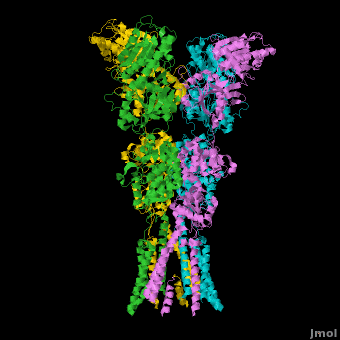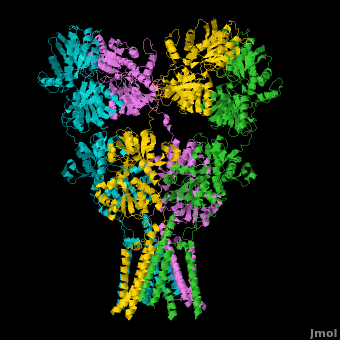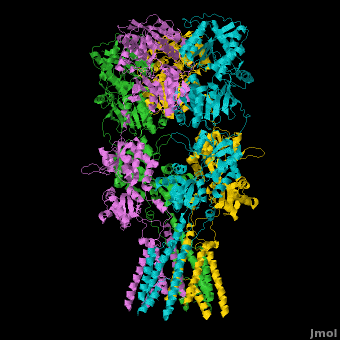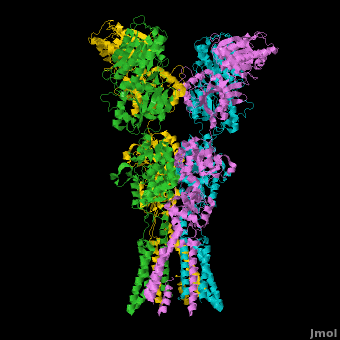Ionotropic Glutamate Receptors
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
AMPA IGluRs form <scene name='Ionotropic_Glutamate_Receptors/Monomer/1'>homotetramers</scene>. Each subunit includes an extracellular <scene name='Ionotropic_Glutamate_Receptors/Atd_opening/1'>amino terminal domain</scene> (ATD) which is responsible for receptor trafficking and modulation, a <scene name='Ionotropic_Glutamate_Receptors/Lbd_opening/1'>ligand-binding domain</scene> (LBD) which activates the receptor upon binding glutamate, and a <scene name='Ionotropic_Glutamate_Receptors/Tmd_opening/1'>transmembrane domain</scene> (TMD) which forms the membrane-spanning ion channel. Also present is a carboxy-terminal domain involved in receptor localization and regulation, although the structure of this domain has not been solved.<ref name="Sobo"/> The structure of AMPA IGluRs or in this case GluA2, is unique in that the <scene name='Ionotropic_Glutamate_Receptors/Sub_a_and_b/1'>symmetry of the receptor changes depending on the domain</scene>. The ATD has a local two-fold symmetry, the LBD has a two-fold symmetry, while the TMD has a four-fold symmetry. Here is a morph depicting the <scene name='Ionotropic_Glutamate_Receptors/Morph_a_to_b/2'>differnce between subunit type A and B</scene>. This symmetry mismatch has implications for function of the receptor with subunits behaving differently depending upon their orientation despite identical primary sequence.<ref name="Sobo"/> For an excellent analysis, see: [[Glutamate_receptor_%28GluA2%29|Glutamate Receptor Symmetry Analysis]] | AMPA IGluRs form <scene name='Ionotropic_Glutamate_Receptors/Monomer/1'>homotetramers</scene>. Each subunit includes an extracellular <scene name='Ionotropic_Glutamate_Receptors/Atd_opening/1'>amino terminal domain</scene> (ATD) which is responsible for receptor trafficking and modulation, a <scene name='Ionotropic_Glutamate_Receptors/Lbd_opening/1'>ligand-binding domain</scene> (LBD) which activates the receptor upon binding glutamate, and a <scene name='Ionotropic_Glutamate_Receptors/Tmd_opening/1'>transmembrane domain</scene> (TMD) which forms the membrane-spanning ion channel. Also present is a carboxy-terminal domain involved in receptor localization and regulation, although the structure of this domain has not been solved.<ref name="Sobo"/> The structure of AMPA IGluRs or in this case GluA2, is unique in that the <scene name='Ionotropic_Glutamate_Receptors/Sub_a_and_b/1'>symmetry of the receptor changes depending on the domain</scene>. The ATD has a local two-fold symmetry, the LBD has a two-fold symmetry, while the TMD has a four-fold symmetry. Here is a morph depicting the <scene name='Ionotropic_Glutamate_Receptors/Morph_a_to_b/2'>differnce between subunit type A and B</scene>. This symmetry mismatch has implications for function of the receptor with subunits behaving differently depending upon their orientation despite identical primary sequence.<ref name="Sobo"/> For an excellent analysis, see: [[Glutamate_receptor_%28GluA2%29|Glutamate Receptor Symmetry Analysis]] | ||
=====The Amino Terminal Domain===== | =====The Amino Terminal Domain===== | ||
| - | | + | <scene name='Ionotropic_Glutamate_Receptors/Atd_opening/2'>The ATD</scene> is responsible for receptor assembly, trafficking and localization. It has two unique sets of interactions which hold the tetramer together. The <scene name='Ionotropic_Glutamate_Receptors/Atd_dimer_interactions/1'>first set of interactions</scene> is present in each pair of dimers and involves both hydrogen bonding and hydrophobic interactions. The **second set**, which includes residues >>>>>>>>>>>>, effectively holds the pair of dimers together at an angle that is roughly 24 degrees off of the overall two-fold axis.<ref name="Sobo"/><ref>PMID: 19461580</ref> |
=====The Transmembrane Domain===== | =====The Transmembrane Domain===== | ||
**The TMD** has a pore structure that is nearly identical to that of the [[Potassium Channel]]. With complete four-fold symmetry, 16 helices form a **precise pore** through which cations can flow through. In the current, inhibitor bound structure, the M3 helices cross at a highly conserved **SYTANLAAF motif**, with Thr 617, Ala 621, and Thr 625 **occluding the ion permeation pathway**.<ref name="Sobo"/> The **narrowest part** of the channel includes the residues Met 629, Thr 625, Ala 621, and Thr 617. Located next to this narrow region lies **Alanine 622**, which is replaced with a threonine in the Lurcher mouse model mentioned previously. This mutation, which introduces a significantly bulkier residue, destabilizes the tight helix crossing associated with the closed state of the receptor, resulting in a constitutively open ion channel.<ref name="Sobo"/> | **The TMD** has a pore structure that is nearly identical to that of the [[Potassium Channel]]. With complete four-fold symmetry, 16 helices form a **precise pore** through which cations can flow through. In the current, inhibitor bound structure, the M3 helices cross at a highly conserved **SYTANLAAF motif**, with Thr 617, Ala 621, and Thr 625 **occluding the ion permeation pathway**.<ref name="Sobo"/> The **narrowest part** of the channel includes the residues Met 629, Thr 625, Ala 621, and Thr 617. Located next to this narrow region lies **Alanine 622**, which is replaced with a threonine in the Lurcher mouse model mentioned previously. This mutation, which introduces a significantly bulkier residue, destabilizes the tight helix crossing associated with the closed state of the receptor, resulting in a constitutively open ion channel.<ref name="Sobo"/> | ||
Revision as of 23:40, 12 March 2011
| |||||||||||
Additional Resources
For additional information on the Symmetry of the Glutamate Receptor, See: Glutamate Receptor Symmetry Analysis
For Additional Information, See: Membrane Channels & Pumps
For Additional Information, See: Alzheimer's Disease
References
- ↑ 1.0 1.1 1.2 Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E, Partin KM. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005 Sep 28;25(39):9027-36. PMID:16192394 doi:25/39/9027
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
- ↑ 3.0 3.1 3.2 3.3 Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001 Nov 13;57(9):1618-28. PMID:11706102
- ↑ Welsh JP, Ahn ES, Placantonakis DG. Is autism due to brain desynchronization? Int J Dev Neurosci. 2005 Apr-May;23(2-3):253-63. PMID:15749250 doi:10.1016/j.ijdevneu.2004.09.002
- ↑ Zuo J, De Jager PL, Takahashi KA, Jiang W, Linden DJ, Heintz N. Neurodegeneration in Lurcher mice caused by mutation in delta2 glutamate receptor gene. Nature. 1997 Aug 21;388(6644):769-73. PMID:9285588 doi:10.1038/42009
- ↑ Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003 Oct;2(5):255-67. PMID:14606691
- ↑ Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009 Jun 17;28(12):1812-23. Epub 2009 May 21. PMID:19461580 doi:10.1038/emboj.2009.140
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Wayne Decatur, Alexander Berchansky, Joel L. Sussman