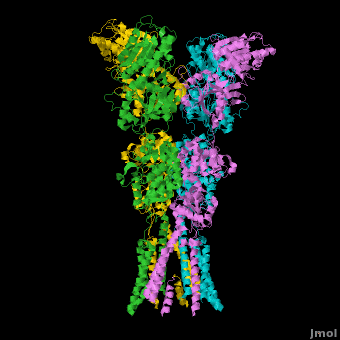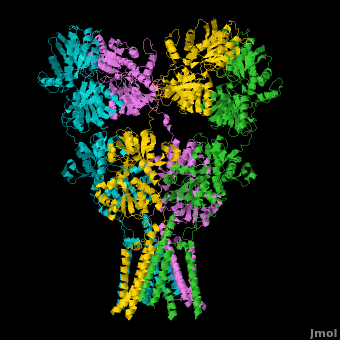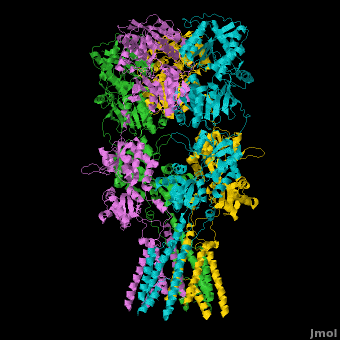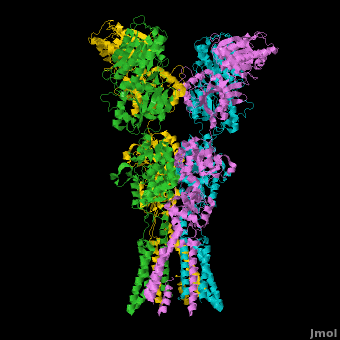Ionotropic Glutamate Receptors
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
AMPA IGluRs form <scene name='Ionotropic_Glutamate_Receptors/Monomer/1'>homotetramers</scene>. Each subunit includes an extracellular <scene name='Ionotropic_Glutamate_Receptors/Atd_opening/1'>amino terminal domain</scene> (ATD) which is responsible for receptor trafficking and modulation, a <scene name='Ionotropic_Glutamate_Receptors/Lbd_opening/1'>ligand-binding domain</scene> (LBD) which activates the receptor upon binding glutamate, and a <scene name='Ionotropic_Glutamate_Receptors/Tmd_opening/1'>transmembrane domain</scene> (TMD) which forms the membrane-spanning ion channel. Also present is a carboxy-terminal domain involved in receptor localization and regulation, although the structure of this domain has not been solved.<ref name="Sobo"/> The structure of AMPA IGluRs or in this case GluA2, is unique in that the <scene name='Ionotropic_Glutamate_Receptors/Sub_a_and_b/1'>symmetry of the receptor changes depending on the domain</scene>. The ATD has a local two-fold symmetry, the LBD has a two-fold symmetry, while the TMD has a four-fold symmetry. Here is a morph depicting the <scene name='Ionotropic_Glutamate_Receptors/Morph_a_to_b/2'>differnce between subunit type A and B</scene>. This symmetry mismatch has implications for function of the receptor with subunits behaving differently depending upon their orientation despite identical primary sequence.<ref name="Sobo"/> For an excellent analysis, see: [[Glutamate_receptor_%28GluA2%29|Glutamate Receptor Symmetry Analysis]] | AMPA IGluRs form <scene name='Ionotropic_Glutamate_Receptors/Monomer/1'>homotetramers</scene>. Each subunit includes an extracellular <scene name='Ionotropic_Glutamate_Receptors/Atd_opening/1'>amino terminal domain</scene> (ATD) which is responsible for receptor trafficking and modulation, a <scene name='Ionotropic_Glutamate_Receptors/Lbd_opening/1'>ligand-binding domain</scene> (LBD) which activates the receptor upon binding glutamate, and a <scene name='Ionotropic_Glutamate_Receptors/Tmd_opening/1'>transmembrane domain</scene> (TMD) which forms the membrane-spanning ion channel. Also present is a carboxy-terminal domain involved in receptor localization and regulation, although the structure of this domain has not been solved.<ref name="Sobo"/> The structure of AMPA IGluRs or in this case GluA2, is unique in that the <scene name='Ionotropic_Glutamate_Receptors/Sub_a_and_b/1'>symmetry of the receptor changes depending on the domain</scene>. The ATD has a local two-fold symmetry, the LBD has a two-fold symmetry, while the TMD has a four-fold symmetry. Here is a morph depicting the <scene name='Ionotropic_Glutamate_Receptors/Morph_a_to_b/2'>differnce between subunit type A and B</scene>. This symmetry mismatch has implications for function of the receptor with subunits behaving differently depending upon their orientation despite identical primary sequence.<ref name="Sobo"/> For an excellent analysis, see: [[Glutamate_receptor_%28GluA2%29|Glutamate Receptor Symmetry Analysis]] | ||
=====The Amino Terminal Domain===== | =====The Amino Terminal Domain===== | ||
| - | <scene name='Ionotropic_Glutamate_Receptors/Atd_opening/2'>The ATD</scene> is responsible for receptor assembly, trafficking and localization. It has two unique sets of interactions which hold the tetramer together. The <scene name='Ionotropic_Glutamate_Receptors/Atd_dimer_interactions/1'>first set of interactions</scene> is present in each pair of dimers and involves both hydrogen bonding and hydrophobic interactions. The | + | <scene name='Ionotropic_Glutamate_Receptors/Atd_opening/2'>The ATD</scene> is responsible for receptor assembly, trafficking and localization. It has two unique sets of interactions which hold the tetramer together. The <scene name='Ionotropic_Glutamate_Receptors/Atd_dimer_interactions/1'>first set of interactions</scene> is present in each pair of dimers and involves both hydrogen bonding and hydrophobic interactions. The <scene name='Ionotropic_Glutamate_Receptors/Atd_two_dimers_interaction/2'>second set</scene>,which includes residues Ile 203, Thr 204, Ile 205, and Val 209 on both chains among others, effectively holds the pair of dimers together at an angle that is roughly 24 degrees off of the overall two-fold axis.<ref name="Sobo"/><ref>PMID: 19461580</ref> |
=====The Transmembrane Domain===== | =====The Transmembrane Domain===== | ||
**The TMD** has a pore structure that is nearly identical to that of the [[Potassium Channel]]. With complete four-fold symmetry, 16 helices form a **precise pore** through which cations can flow through. In the current, inhibitor bound structure, the M3 helices cross at a highly conserved **SYTANLAAF motif**, with Thr 617, Ala 621, and Thr 625 **occluding the ion permeation pathway**.<ref name="Sobo"/> The **narrowest part** of the channel includes the residues Met 629, Thr 625, Ala 621, and Thr 617. Located next to this narrow region lies **Alanine 622**, which is replaced with a threonine in the Lurcher mouse model mentioned previously. This mutation, which introduces a significantly bulkier residue, destabilizes the tight helix crossing associated with the closed state of the receptor, resulting in a constitutively open ion channel.<ref name="Sobo"/> | **The TMD** has a pore structure that is nearly identical to that of the [[Potassium Channel]]. With complete four-fold symmetry, 16 helices form a **precise pore** through which cations can flow through. In the current, inhibitor bound structure, the M3 helices cross at a highly conserved **SYTANLAAF motif**, with Thr 617, Ala 621, and Thr 625 **occluding the ion permeation pathway**.<ref name="Sobo"/> The **narrowest part** of the channel includes the residues Met 629, Thr 625, Ala 621, and Thr 617. Located next to this narrow region lies **Alanine 622**, which is replaced with a threonine in the Lurcher mouse model mentioned previously. This mutation, which introduces a significantly bulkier residue, destabilizes the tight helix crossing associated with the closed state of the receptor, resulting in a constitutively open ion channel.<ref name="Sobo"/> | ||
Revision as of 23:53, 12 March 2011
| |||||||||||
Additional Resources
For additional information on the Symmetry of the Glutamate Receptor, See: Glutamate Receptor Symmetry Analysis
For Additional Information, See: Membrane Channels & Pumps
For Additional Information, See: Alzheimer's Disease
References
- ↑ 1.0 1.1 1.2 Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E, Partin KM. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005 Sep 28;25(39):9027-36. PMID:16192394 doi:25/39/9027
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
- ↑ 3.0 3.1 3.2 3.3 Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001 Nov 13;57(9):1618-28. PMID:11706102
- ↑ Welsh JP, Ahn ES, Placantonakis DG. Is autism due to brain desynchronization? Int J Dev Neurosci. 2005 Apr-May;23(2-3):253-63. PMID:15749250 doi:10.1016/j.ijdevneu.2004.09.002
- ↑ Zuo J, De Jager PL, Takahashi KA, Jiang W, Linden DJ, Heintz N. Neurodegeneration in Lurcher mice caused by mutation in delta2 glutamate receptor gene. Nature. 1997 Aug 21;388(6644):769-73. PMID:9285588 doi:10.1038/42009
- ↑ Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003 Oct;2(5):255-67. PMID:14606691
- ↑ Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009 Jun 17;28(12):1812-23. Epub 2009 May 21. PMID:19461580 doi:10.1038/emboj.2009.140
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Wayne Decatur, Alexander Berchansky, Joel L. Sussman