Sandbox Reserved 197
From Proteopedia
| Line 1: | Line 1: | ||
| - | <!-- PLEASE DO NOT DELETE THIS TEMPLATE --> | + | <Structure load='7rsa' size='500' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /><!-- PLEASE DO NOT DELETE THIS TEMPLATE --> |
{{Template:Johnson_CH462_Spring2011}} | {{Template:Johnson_CH462_Spring2011}} | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
Revision as of 22:01, 29 March 2011
|
| This Sandbox is Reserved from Feb 02, 2011, through Jul 31, 2011 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 191 through Sandbox Reserved 200. |
To get started:
More help: Help:Editing |
Introduction
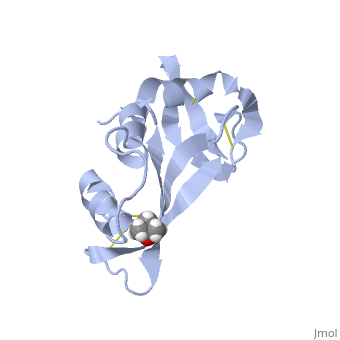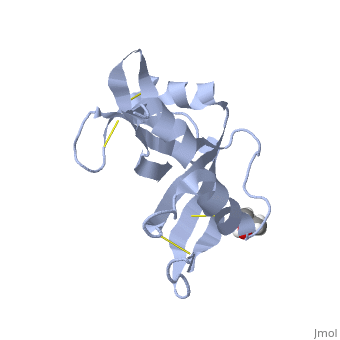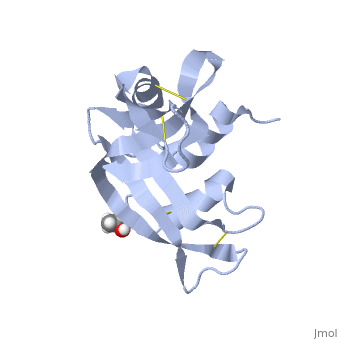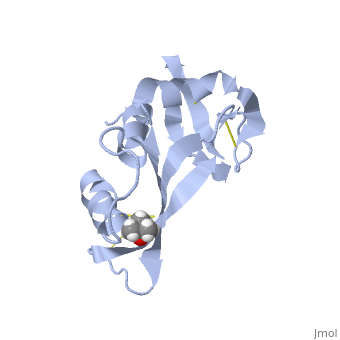
Ribonuclease A is an enzyme involved in catalyzing RNA degradation. The structure of RNase A has been determined through crystallization and FABMS. There are several features of its structure that are pertinent to its folding and function and RNase A has been used time and again to illustrate these important features of protein folding. The active siteof a protein is where the actually binding of the protein to its substrate(s) occur(s) and the active site of Rnase A lies within its cleft. Proteins form interactions between different parts of their structure for stabilization. In RNase A, there are eight cysteine residues present that form four disulfide linkages that contribute to the ordered structure as well as the speed of folding of RNase A. Another important aspect to folding of its is the presence of two cis proline residues.
Folding
There are features of every protein that directly or indirectly effect the folding of that protein. Several of these features have been identified in RNase A by the use of mutants of the native form. These mutations and the study of the kinetics and final structure in comparison to the native form show whether that particular feature is involved in the folding of the protein. One particular feature of RNase A is the presence of cis proline residues. In nature, most amino acids reside in a trans conformation. Due to their cyclic structure, prolines are more stable in a cis conformation. RNase A contains four proline residues, two reside in the "cis" conformation and two in the "trans" conformation. The peptide group of RNase A in its native state is found in the cis conformation. Despite a mutation, a "cis" conformation still forms; this is an unlikely conformation for an alanine residue. Upon unfolding, Tyr92-Ala93 undergoes isomerization to form its favored "trans" conformation. This points to the fact that this "cis" bond formation is a key component to the protein structure of RNase A.
Another important feature of the folding of RNase A is the presence of four disulfide bonds. These bonds contribute to the thermal stability and the rate of folding of RNase A. The residues involved in these linkages include , , , and . Cys26-Cys84 and Cys58-Cys110 create an interaction between an alpha-helix and a beta sheet. This connection is the main contributor to the thermodynamic stability. RNase A actually has a rate-determining three-disulfide intermediate. An analog of this, , shows RNase A, missing the disulfide bond that would normally occur here, . As you can see in the variant, there are only 3 disulfide bonds present, shown in blue.