Large Ribosomal Subunit of Haloarcula
From Proteopedia
m (→The ribosome is a ribozyme - protein DOES NOT participate directly in the chemistry of peptide bond synthesis:) |
|||
| Line 3: | Line 3: | ||
|OLDID=1108565 | |OLDID=1108565 | ||
}} | }} | ||
| - | [[ | + | |
| + | <StructureSection load='1s72simplified.PDB.gz' size='450' frame='true' align='right' scene='User:Wayne_Decatur/Sandbox_Haloarcula_Ribosomal_Large_Subunit/1s72/6' >'''The Large Ribosomal Subunit''' ([[1s72]]), resolution 2.4Å (<scene name='User:Wayne_Decatur/Sandbox_Haloarcula_Ribosomal_Large_Subunit/1s72/6'>initial scene</scene>). <br> | ||
| + | ·· {{Link Toggle halolsuriborRNA}} ·· {{Link Toggle halolsu23SrRNA}} ·· {{Link Toggle halolsu5SrRNA}} ··<br>·· {{Link Toggle 70SriboProtein}} ·· {{Link Toggle BlackWhiteBackground}} | ||
'''The ''Haloarcula'' Large Ribosomal Subunit'''<br> | '''The ''Haloarcula'' Large Ribosomal Subunit'''<br> | ||
The molecular machine catalyzing petide bond synthesis is a ribozyme. | The molecular machine catalyzing petide bond synthesis is a ribozyme. | ||
| - | |||
| - | |||
| - | |||
| - | |||
==Introduction== | ==Introduction== | ||
The [[ribosome]] is a complex composed of RNA and protein that adds up to several million daltons in size and plays a critical role in the process of decoding the genetic information stored in the genome into protein as outlined in what is now known as the Central Dogma of Molecular Biology<ref>[http://sandwalk.blogspot.com/2009/10/ribosome-and-central-dogma-of-molecular.html the Central Dogma of Molecular Biology clarified]</ref>. Specifically, the ribosome carries out the process of translation, decoding the genetic information encoded in messenger RNA, one amino acid at a time, into newly synthesized polypeptide chains. The ribosome functions as a complex of two complexes of many proteins and RNAs of substantial length; these two complexes are the small ribosomal subunit and the large ribosomal subunit. The formation of peptide bonds occurs in the large subunit where the acceptor-stems of the tRNAs are docked. | The [[ribosome]] is a complex composed of RNA and protein that adds up to several million daltons in size and plays a critical role in the process of decoding the genetic information stored in the genome into protein as outlined in what is now known as the Central Dogma of Molecular Biology<ref>[http://sandwalk.blogspot.com/2009/10/ribosome-and-central-dogma-of-molecular.html the Central Dogma of Molecular Biology clarified]</ref>. Specifically, the ribosome carries out the process of translation, decoding the genetic information encoded in messenger RNA, one amino acid at a time, into newly synthesized polypeptide chains. The ribosome functions as a complex of two complexes of many proteins and RNAs of substantial length; these two complexes are the small ribosomal subunit and the large ribosomal subunit. The formation of peptide bonds occurs in the large subunit where the acceptor-stems of the tRNAs are docked. | ||
| Line 28: | Line 26: | ||
==Haloracula Large Ribosomal Subunit Components== | ==Haloracula Large Ribosomal Subunit Components== | ||
| - | + | ||
| - | + | ||
The large subunit of the <em>Haloracula marismortui</em> ribosome sediments at 50S, as do the large subunits of archaea and eubacteria. It is composed of two chains of RNA, a 23S chain (2,922 nucleotides long, 946 kDa) and a 5S chain (122 bases long, 39 kDa). Assembled with the RNA are 27 protein chains (of a total of 31 known), varying in length from 49 (L39E, 6 kDa) to 337 amino acids (L3, 37 kDa)<ref>PMID:10937989</ref>. | The large subunit of the <em>Haloracula marismortui</em> ribosome sediments at 50S, as do the large subunits of archaea and eubacteria. It is composed of two chains of RNA, a 23S chain (2,922 nucleotides long, 946 kDa) and a 5S chain (122 bases long, 39 kDa). Assembled with the RNA are 27 protein chains (of a total of 31 known), varying in length from 49 (L39E, 6 kDa) to 337 amino acids (L3, 37 kDa)<ref>PMID:10937989</ref>. | ||
| Line 51: | Line 48: | ||
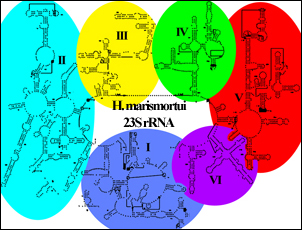
The secondary structure map of Haloarcula 23S rRNA (below) clearly shows six large RNA domains extending off a large major loop.<br> | The secondary structure map of Haloarcula 23S rRNA (below) clearly shows six large RNA domains extending off a large major loop.<br> | ||
[[Image:Schematic hmarlsu.jpg|left]] | [[Image:Schematic hmarlsu.jpg|left]] | ||
| - | + | ||
| - | + | ||
| Line 85: | Line 81: | ||
===The proteins:=== | ===The proteins:=== | ||
| - | + | ||
| - | + | ||
:<scene name='User:Wayne_Decatur/Sandbox_Haloarcula_Ribosomal_Large_Subunit/1s72extenvsglb/6'>Globular vs. extended proteins</scene> | :<scene name='User:Wayne_Decatur/Sandbox_Haloarcula_Ribosomal_Large_Subunit/1s72extenvsglb/6'>Globular vs. extended proteins</scene> | ||
:*Proteins are generally globular. However, while about half the 27 proteins seen in the crystal structure of the large ribosomal subunit are globular (<font color="orange">'''shown as orange'''</font>), interestingly, '''the other half are extended or have large extended regions''' emanating from globular domains (<font color="cyan">'''shown as cyan'''</font>). | :*Proteins are generally globular. However, while about half the 27 proteins seen in the crystal structure of the large ribosomal subunit are globular (<font color="orange">'''shown as orange'''</font>), interestingly, '''the other half are extended or have large extended regions''' emanating from globular domains (<font color="cyan">'''shown as cyan'''</font>). | ||
| Line 123: | Line 118: | ||
===The ribosome is a ribozyme - protein DOES NOT participate directly in the chemistry of peptide bond synthesis:=== | ===The ribosome is a ribozyme - protein DOES NOT participate directly in the chemistry of peptide bond synthesis:=== | ||
| - | + | ||
| - | + | ||
:<scene name='User:Wayne_Decatur/Sandbox_Haloarcula_Ribosomal_Large_Subunit/1s72rrnadomain5allwithyarus/9'>An informative analog is observed at the core of the large subunit bound to Domain V</scene> (<font color = "red">'''shown in red'''</font>) | :<scene name='User:Wayne_Decatur/Sandbox_Haloarcula_Ribosomal_Large_Subunit/1s72rrnadomain5allwithyarus/9'>An informative analog is observed at the core of the large subunit bound to Domain V</scene> (<font color = "red">'''shown in red'''</font>) | ||
:*In addition to unliganded subunit, the large subunit structure has been solved with substrate analogs which provides a detailed view of the direct role <font color="red">'''domain V'''</font> plays in the chemistry of peptide bond synthesis. One of the analogs was the <font color="magenta">'''Yarus analog (CCA-puromycin)'''</font>, known to inhibit translation because it mimics normal substrate, specifically it resembles an unstable transition state intermediate formed during peptide bond synthesis and involving the extreme ends of the A- and P- tRNAs and atoms corresponding to parts of an amino acid. <scene name='User:Wayne_Decatur/Sandbox_Haloarcula_Ribosomal_Large_Subunit/1s72rrnadomain5withyarus/4'>The Yarus analog (magenta), indicating the site of the petidyl transferase reaction, is entrenched in Domain V</scene>. | :*In addition to unliganded subunit, the large subunit structure has been solved with substrate analogs which provides a detailed view of the direct role <font color="red">'''domain V'''</font> plays in the chemistry of peptide bond synthesis. One of the analogs was the <font color="magenta">'''Yarus analog (CCA-puromycin)'''</font>, known to inhibit translation because it mimics normal substrate, specifically it resembles an unstable transition state intermediate formed during peptide bond synthesis and involving the extreme ends of the A- and P- tRNAs and atoms corresponding to parts of an amino acid. <scene name='User:Wayne_Decatur/Sandbox_Haloarcula_Ribosomal_Large_Subunit/1s72rrnadomain5withyarus/4'>The Yarus analog (magenta), indicating the site of the petidyl transferase reaction, is entrenched in Domain V</scene>. | ||
| Line 138: | Line 132: | ||
===Polypeptide Exit Tunnel:=== | ===Polypeptide Exit Tunnel:=== | ||
| - | + | ||
| - | + | ||
:<scene name='User:Wayne_Decatur/Sandbox_Haloarcula_Ribosomal_Large_Subunit/1s72tunnelend/2'>The end of the Polypeptide Exit Tunnel is shown in the center</scene> | :<scene name='User:Wayne_Decatur/Sandbox_Haloarcula_Ribosomal_Large_Subunit/1s72tunnelend/2'>The end of the Polypeptide Exit Tunnel is shown in the center</scene> | ||
:*As the nascent chain grows, it advances into <font color="dodgerblue">'''a tunnel'''</font> about 100 angstroms long that passes through the large subunit, called the polypeptide exit tunnel. | :*As the nascent chain grows, it advances into <font color="dodgerblue">'''a tunnel'''</font> about 100 angstroms long that passes through the large subunit, called the polypeptide exit tunnel. | ||
| Line 171: | Line 164: | ||
* For additional information, see: [[Bacterial Infections]] | * For additional information, see: [[Bacterial Infections]] | ||
* For additional information, see: [[Translation]] | * For additional information, see: [[Translation]] | ||
| - | + | </StructureSection> | |
==References and Notes== | ==References and Notes== | ||
<references /> | <references /> | ||
Revision as of 14:44, 20 March 2013
This page, as it appeared on August 1, 2010, was featured in this article in the journal Biochemistry and Molecular Biology Education.
| |||||||||||
References and Notes
- ↑ the Central Dogma of Molecular Biology clarified
- ↑ The Nobel Prize in Chemistry 2009 page at The Official Web Site of the Nobel Prize
- ↑ Steitz TA. From the structure and function of the ribosome to new antibiotics (Nobel Lecture). Angew Chem Int Ed Engl. 2010 Jun 14;49(26):4381-98. PMID:20509130 doi:10.1002/anie.201000708
- ↑ Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000 Aug 11;289(5481):905-20. PMID:10937989
- ↑ Ban N, Freeborn B, Nissen P, Penczek P, Grassucci RA, Sweet R, Frank J, Moore PB, Steitz TA. A 9 A resolution X-ray crystallographic map of the large ribosomal subunit. Cell. 1998 Jun 26;93(7):1105-15. PMID:9657144
- ↑ Helix 69 is shown clearly in Domain IV in a detailed secondary structure of Haloarcula marismortui 23S rRNA
- ↑ Gao N, Zavialov AV, Ehrenberg M, Frank J. Specific interaction between EF-G and RRF and its implication for GTP-dependent ribosome splitting into subunits. J Mol Biol. 2007 Dec 14;374(5):1345-58. Epub 2007 Oct 16. PMID:17996252 doi:10.1016/j.jmb.2007.10.021
- ↑ Kipper K, Hetenyi C, Sild S, Remme J, Liiv A. Ribosomal intersubunit bridge B2a is involved in factor-dependent translation initiation and translational processivity. J Mol Biol. 2009 Jan 16;385(2):405-22. Epub 2008 Nov 5. PMID:19007789 doi:10.1016/j.jmb.2008.10.065
- ↑ Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell. 2006 Sep 15;23(6):865-74. PMID:16973438 doi:10.1016/j.molcel.2006.08.011
- ↑ Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000 Aug 11;289(5481):920-30. PMID:10937990
- ↑ Maguire BA, Beniaminov AD, Ramu H, Mankin AS, Zimmermann RA. A protein component at the heart of an RNA machine: the importance of protein l27 for the function of the bacterial ribosome. Mol Cell. 2005 Nov 11;20(3):427-35. PMID:16285924 doi:10.1016/j.molcel.2005.09.009
- ↑ Trobro S, Aqvist J. Role of ribosomal protein L27 in peptidyl transfer. Biochemistry. 2008 Apr 29;47(17):4898-906. Epub 2008 Apr 8. PMID:18393533 doi:10.1021/bi8001874
- ↑ Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009 May;16(5):528-33. Epub 2009 Apr 12. PMID:19363482 doi:10.1038/nsmb.1577
- ↑ Voss NR, Gerstein M, Steitz TA, Moore PB. The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol. 2006 Jul 21;360(4):893-906. Epub 2006 May 30. PMID:16784753 doi:10.1016/j.jmb.2006.05.023
- ↑ Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002 Mar 8;108(5):629-36. PMID:11893334
- ↑ Berisio R, Schluenzen F, Harms J, Bashan A, Auerbach T, Baram D, Yonath A. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat Struct Biol. 2003 May;10(5):366-70. PMID:12665853 doi:10.1038/nsb915
- ↑ Gabashvili IS, Gregory ST, Valle M, Grassucci R, Worbs M, Wahl MC, Dahlberg AE, Frank J. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol Cell. 2001 Jul;8(1):181-8. PMID:11511371
- ↑ Lawrence MG, Lindahl L, Zengel JM. Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J Bacteriol. 2008 Sep;190(17):5862-9. Epub 2008 Jun 27. PMID:18586934 doi:10.1128/JB.00632-08
- ↑ Fulle S, Gohlke H. Statics of the ribosomal exit tunnel: implications for cotranslational peptide folding, elongation regulation, and antibiotics binding. J Mol Biol. 2009 Mar 27;387(2):502-17. Epub 2009 Jan 27. PMID:19356596 doi:10.1016/j.jmb.2009.01.037
- ↑ Zengel JM, Jerauld A, Walker A, Wahl MC, Lindahl L. The extended loops of ribosomal proteins L4 and L22 are not required for ribosome assembly or L4-mediated autogenous control. RNA. 2003 Oct;9(10):1188-97. PMID:13130133
- ↑ Giglione C, Fieulaine S, Meinnel T. Cotranslational processing mechanisms: towards a dynamic 3D model. Trends Biochem Sci. 2009 Aug;34(8):417-26. Epub 2009 Jul 31. PMID:19647435 doi:10.1016/j.tibs.2009.04.003
- ↑ Pool MR. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase. J Cell Biol. 2009 Jun 1;185(5):889-902. Epub 2009 May 25. PMID:19468070 doi:10.1083/jcb.200807066
- ↑ Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000 Aug 11;289(5481):905-20. PMID:10937989
- ↑ Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000 Aug 11;289(5481):920-30. PMID:10937990
- ↑ Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001 Aug 1;20(15):4214-21. PMID:11483524 doi:http://dx.doi.org/10.1093/emboj/20.15.4214
- ↑ Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004 Jun 25;340(1):141-77. PMID:15184028 doi:10.1016/j.jmb.2004.03.076
- ↑ Kavran JM, Steitz TA. Structure of the base of the L7/L12 stalk of the Haloarcula marismortui large ribosomal subunit: analysis of L11 movements. J Mol Biol. 2007 Aug 24;371(4):1047-59. Epub 2007 Jun 4. PMID:17599351 doi:10.1016/j.jmb.2007.05.091
- ↑ Blaha G, Gurel G, Schroeder SJ, Moore PB, Steitz TA. Mutations outside the anisomycin-binding site can make ribosomes drug-resistant. J Mol Biol. 2008 Jun 6;379(3):505-19. Epub 2008 Apr 8. PMID:18455733 doi:http://dx.doi.org/10.1016/j.jmb.2008.03.075
- ↑ Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000 Aug 11;289(5481):905-20. PMID:10937989
Additional Literature
- Moore PB. The ribosome returned. J Biol. 2009;8(1):8. Epub 2009 Jan 26. PMID:19222865 doi:10.1186/jbiol103
- Steitz TA. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol. 2008 Mar;9(3):242-53. PMID:18292779 doi:10.1038/nrm2352
- Rodnina MV, Wintermeyer W. The ribosome goes Nobel. Trends Biochem Sci. 2010 Jan;35(1):1-5. Epub 2009 Dec 2. PMID:19962317 doi:10.1016/j.tibs.2009.11.003
- Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009 Oct 29;461(7268):1234-42. Epub 2009 Oct 18. PMID:19838167 doi:10.1038/nature08403
- Ramakrishnan V, Moore PB. Atomic structures at last: the ribosome in 2000. Curr Opin Struct Biol. 2001 Apr;11(2):144-54. PMID:11297922
- Schroeder KT, McPhee SA, Ouellet J, Lilley DM. A structural database for k-turn motifs in RNA. RNA. 2010 Aug;16(8):1463-8. Epub 2010 Jun 18. PMID:20562215 doi:10.1261/rna.2207910
External Resources
- 70S Ribosome: January 2010 Molecule of the Month as part of the series of tutorials that are at the RCSB Protein Data Bank and written by David Goodsell
- RCSB Protein Data Bank coverage of the 2009 Nobel Prizes in Chemistry
- The Nobel Prize in Chemistry 2009 page at The Official Web Site of the Nobel Prize
- Ribosome: October 2000 Molecule of the Month as part of the series of tutorials that are at the RCSB Protein Data Bank and written by David Goodsell
- A detailed secondary structure of Haloarcula marismortui 23S rRNA at the 3D Ribosomal Modifications Map Database
Proteopedia Page Contributors and Editors (what is this?)
Wayne Decatur, Michal Harel, Joel L. Sussman, David Canner, Eran Hodis, Angel Herraez, Alexander Berchansky
Categories: Haloarcula marismortui | Klein, D J. | Moore, P B. | Schmeing, T M. | Steitz, T A. | Ban, N. | Hansen, J. | Nissen, P. | Blaha, G. | Gurel, G. | Schroeder, S J. | Genomic sequnece for r-protein | Metal-binding | Protein-protein | Protein-rna | Ribosome assembly | Tunnel | Ribozyme | Intrinsically Unfolded Proteins | Antibiotic | Puromycin | Macrolide | Azithromycin | Zithromax | TRNA | Rna-rna | 50S | Large ribosomal subunit | Ribosome | Translation | Peptide bond formation | Kink-turn | K-turn | A-minor motif | Ribose zipper | Acetylation | Rna-binding | Rrna-binding | Trna-binding | Zinc | Zinc-finger | RNA | Topic Page | Featured in BAMBED