User:Bianca Varney/Bacterial Replication Termination
From Proteopedia
| Line 25: | Line 25: | ||
====Tus structure==== | ====Tus structure==== | ||
| - | Tus is a member of the replication termination protein family and has no similarity to any DNA-binding motif that is known, and is organized into two discontinuous domains (N terminal and C terminal) that consist of α helical and β sheets. Two pairs of antiparallel β strands that form an interdomain connect the β sheet strands of the two domains providing a large central cleft that is positively charged into which the double helix (deformed locally) can fit, such that the two α helical domains flank the DNA. The interdomain β strands, which makes up the core region of the 13bp DNA-binding, that sit across from the DNA, accesses a deepened major groove and contacts several of the bases within this groove, and is responsible for ''ter'' sequence recognition and binding to DNA. There are 17 sequence-specific interactions between the DNA and the α helixes and β strands presented by Tus, although the majority of these interactions are from the proximinal β sheets. The helicase-blocking or | + | Tus is a member of the replication termination protein family and has no similarity to any DNA-binding motif that is known, and is organized into two discontinuous domains (N terminal and C terminal) that consist of α helical and β sheets. Two pairs of antiparallel β strands that form an interdomain connect the β sheet strands of the two domains providing a large central cleft that is positively charged into which the double helix (deformed locally) can fit, such that the two α helical domains flank the DNA. The interdomain β strands, which makes up the core region of the 13bp DNA-binding, that sit across from the DNA, accesses a deepened major groove and contacts several of the bases within this groove, and is responsible for ''ter'' sequence recognition and binding to DNA. There are 17 sequence-specific interactions between the DNA and the α helixes and β strands presented by Tus, although the majority of these interactions are from the proximinal β sheets. The helicase-blocking or , of Tus consists of α helices and loops from N and C terminal domains. Tus is unrelated structurally to the replication termination protein despite their similar functions. |
====Tus Mechanism of Action==== | ====Tus Mechanism of Action==== | ||
Revision as of 03:02, 21 May 2011
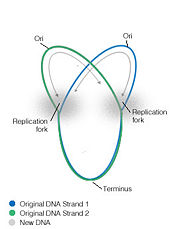
In most bacterial DNA replication initiation occurs at an origin where, due to the circular nature of the chromosome, the replication forks move bidirectionally to end at approxiametly 180 degrees away, at a specific sequence termini region [1]. Bacterial replication termination systems have been well studied in Eschericia coli and Bascillus subtilis. In both systems a trans-acting replication termination protein binds to a specific cis-acting DNA sequences, the replication termini (ter), and the DNA-protein complex arrests the progression of replication forks. The terminator sites are orientated so that protein binding is asymmetric, allowing the complexes to block the replication machinery from only one direction while letting them proceed unimpeded from the other direction [2]. In this way they are said to act in a polar manner. The proteins involved in this termination are non-homologous and differ structurally in E.coli and B.subtilis, although each contains similar contrahelicase activity and performs similar functions in arresting replication.
Termination (ter) Sites
Replication is terminated in bacterial systems such as E.coli and B.subtilis by a "replication fork trap", studded with termination sites which causes the bidirectional forks to pause, encounter and fuse within a region called the terminus region. In E.coli the termination regions are spread across nearly half the chromosome compared to B.subtilis where they cover only ~10%. Termination regions are made up of two groups, opposite to each other, containing inverted sequences for the polar arrest of the replication helicase. In E.coli the 5 ter sites, J, G, F, B and C are arranged opposed to ter sites H, I, E, D and A, and can arrest the fork progressing in the clockwise direction and can block the anticlockwise direction, respectively. The replication fork progressing in a clockwise direction will encounter the terC site first and pause. If the fork progressing from the anticlockwise direction meets the clockwise fork while paused, replication is terminated, however if it does not meet its anti-fork it will proceed until it reaches the next termination site, terB, where it will pause again, etc [8]. Therefore multiple ter sites are important as infrequently utilized backups, to ensure that the fork does not leave the terminus region, and that termination is completed. Multiple regions to entrap the replication fork means that if an inactivating mutation arises within a ter site, then arrest can still occur at another ter sequence [6].
Replication Terminator Protein (Bacillus subtilis)
| |||||||||||
The Terminus Utilization Substance (Tus)
| |||||||||||