This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Bacterial Replication Termination
From Proteopedia
(New page: In most bacterial DNA replication initiation occurs at an origin where, due to the circular nature of the chromosome, the replication forks move bidirectionally to end at approxiametly 180...) |
|||
| Line 38: | Line 38: | ||
==Biological Significance== | ==Biological Significance== | ||
| - | The role of the replication fork arrest was primarily believed to be of great importance for the faithful termination of replication, segregation of chromosomes and | + | The role of the replication fork arrest was primarily believed to be of great importance for the faithful termination of replication, segregation of chromosomes and inheritance of a stable genome. However recent studies where the ''rtp'' and ''tus'' genes of ''B.subtilis'' and ''E.coli'', respectively, were knocked out, suggested that this role is dispensable. Indeed, bacterial systems that have mutations within these genes can survive in the environment and appear identical in both growth rate and cell morphology compared to wildtype bacteria, suggesting that replication termination is not a requirement for cellular replication [4]. However as it has analogous presence between different bacteria this suggests that this mechanism is beneficial for these prokaryotes. It has recently been suggested that this form of termination may have roles including; aiding the co-ordination and optimization of recombination events preceding replication; preventing over-replication and preventing the harmful affects of clashes that can occur between replication-transcription due to the bacterial bias in gene orientation. |
| + | |||
| + | As most genes are orientated towards the terminus, from the origin, if replication is not arrested, it progresses into regions being actively transcribed and collides into the transcription RNA polymerase [4]. It is also suggested that termination may occur by specific dif sites, conserved sites that are located near the terminus region that are involved in homologous recombination. In fact the dif-terminus hypothesis proposes that termination occurs at or near these sites, where after termination of the replication forks, the two recombinases, XerC and XerD (proteins originating from E.coli), cause site-specific recombination at these dif-sites, and that this would resolve the catenated chromosomes and complete replication [7]. This mechanism implies that this replication termination by RTP and Tus proteins is merely advantageous to the bacteria and not necessary. | ||
| + | |||
==References== | ==References== | ||
Revision as of 05:52, 21 May 2011
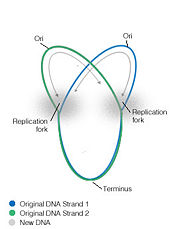
In most bacterial DNA replication initiation occurs at an origin where, due to the circular nature of the chromosome, the replication forks move bidirectionally to end at approxiametly 180 degrees away, at a specific sequence termini region [1]. Bacterial replication termination systems have been well studied in Eschericia coli and Bascillus subtilis. In both systems a trans-acting replication termination protein binds to a specific cis-acting DNA sequences, the replication termini (ter), and the DNA-protein complex arrests the progression of replication forks. The terminator sites are orientated so that protein binding is asymmetric, allowing the complexes to block the replication machinery from only one direction while letting them proceed unimpeded from the other direction [2]. In this way they are said to act in a polar manner. The proteins involved in this termination are non-homologous and differ structurally in E.coli and B.subtilis, although each contains similar contrahelicase activity and performs similar functions in arresting replication.
Contents |
Termination (ter) Sites
Replication is terminated in bacterial systems such as E.coli and B.subtilis by a "replication fork trap", studded with termination sites which causes the bidirectional forks to pause, encounter and fuse within a region called the terminus region. In E.coli the termination regions are spread across nearly half the chromosome compared to B.subtilis where they cover only ~10%. Termination regions are made up of two groups, opposite to each other, containing inverted sequences for the polar arrest of the replication helicase. In E.coli the 5 ter sites, J, G, F, B and C are arranged opposed to ter sites H, I, E, D and A, and can arrest the fork progressing in the clockwise direction and can block the anticlockwise direction, respectively. The replication fork progressing in a clockwise direction will encounter the terC site first and pause. If the fork progressing from the anticlockwise direction meets the clockwise fork while paused, replication is terminated, however if it does not meet its anti-fork it will proceed until it reaches the next termination site, terB, where it will pause again, etc [8]. Therefore multiple ter sites are important as infrequently utilized backups, to ensure that the fork does not leave the terminus region, and that termination is completed. Multiple regions to entrap the replication fork means that if an inactivating mutation arises within a ter site, then arrest can still occur at another ter sequence [6].
Replication Terminator Protein (Bacillus subtilis)
| |||||||||||
The Terminus Utilization Substance (Escherichia coli )
| |||||||||||
Biological Significance
The role of the replication fork arrest was primarily believed to be of great importance for the faithful termination of replication, segregation of chromosomes and inheritance of a stable genome. However recent studies where the rtp and tus genes of B.subtilis and E.coli, respectively, were knocked out, suggested that this role is dispensable. Indeed, bacterial systems that have mutations within these genes can survive in the environment and appear identical in both growth rate and cell morphology compared to wildtype bacteria, suggesting that replication termination is not a requirement for cellular replication [4]. However as it has analogous presence between different bacteria this suggests that this mechanism is beneficial for these prokaryotes. It has recently been suggested that this form of termination may have roles including; aiding the co-ordination and optimization of recombination events preceding replication; preventing over-replication and preventing the harmful affects of clashes that can occur between replication-transcription due to the bacterial bias in gene orientation.
As most genes are orientated towards the terminus, from the origin, if replication is not arrested, it progresses into regions being actively transcribed and collides into the transcription RNA polymerase [4]. It is also suggested that termination may occur by specific dif sites, conserved sites that are located near the terminus region that are involved in homologous recombination. In fact the dif-terminus hypothesis proposes that termination occurs at or near these sites, where after termination of the replication forks, the two recombinases, XerC and XerD (proteins originating from E.coli), cause site-specific recombination at these dif-sites, and that this would resolve the catenated chromosomes and complete replication [7]. This mechanism implies that this replication termination by RTP and Tus proteins is merely advantageous to the bacteria and not necessary.
References
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Django J. Nathan, Michal Harel, Bianca Varney