Structural linkage between ligand discrimination and receptor activation by type I interferons
Christoph Thomas, Ignacio Moraga, Doron Levin, Peter O. Krutzik, Yulia Podoplelova, Angelica Trejo, Choongho Lee, Ganit Yarden, Susan E. Vleck, Jeffrey S. Glenn, Garry P. Nolan, Jacob Piehler, Gideon Schreiber, K. Christopher Garcia[1]
Introduction
IFNs were the first cytokines discovered more than half a century ago as agents that interfere with viral infection. Since then, IFNs have been established as pleiotropic, multifunctional proteins in the early immune response, exhibiting pronounced antiproliferative effects on cells, in addition to their strong immunomodulatory and antiviral activities. Due to their potency and diverse biological activities, IFNs are used for the treatment of several human diseases, including hepatitis C, multiple sclerosis and certain types of cancer.
All initiate signaling by binding to the same cell surface receptor composed of two subunits called and . The intracellular domains (ICDs) of IFNAR1 and IFNAR2 are associated with the Janus kinases (Jaks) Tyk2 and Jak1, respectively. Upon ligand binding by the IFNAR chains and formation of the signaling complex, these tyrosine kinases trans-phosphorylate and thereby activate each other. Subsequently, the activated Jaks phosphorylate STAT transcription factors, which translocate into the nucleus and activate the expression of hundreds of IFN-stimulated genes. To gain insight into how type I IFNs engage their receptor chains, how the receptor system is able to recognize the large number of different ligands, and how different IFN ligands can evoke different physiological activities, we determined the crystal structures of unliganded , the binary complex , and the ternary ligand-receptor complexes of and binding both receptor chains. A final theoretical ternary structure including was also created. These structures, in conjunction with biochemical and cellular experiments, reveal that the type I IFN receptor uses a mode of ligand interaction that is unique among cytokine receptors, but conserved between different IFNs. Furthermore, ligand discrimination occurs through distinct energetics of shared receptor contacts, and differential IFN signaling is mediated by specific ligand-receptor interface chemistries that lead to different ternary complex stabilities.
Interactions Between IFNAR & IFN
A superposition of the two ternary complexes reveals that they have very similar overall architectures, despite the different physiological activities of the IFN ligands. This suggests that the activity differences are not due to different signaling complex architectures. The functional differences are rather mediated by specific interface chemistries that form the basis for different ternary complex stabilities.
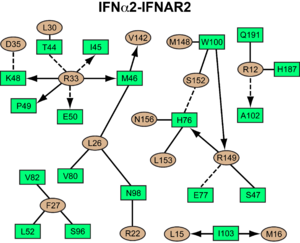
IFNAR2-IFN interaction
interacts primarily with the . Arg33(IFN) appears to be the for the interaction of the IFN ligand with IFNAR2. It forms an with the main chain carbonyl oxygen atoms of . This residue is present in IFNa, IFNw, IFNb and IFNe. Two hydrophobic interaction clusters are part of the interface: is formed between Leu15 and Met16 of the IFN molecule and Trp100 and Ile103 of IFNAR2; comprises Leu26, Phe27, Leu30 and Val142 of the ligand and Met46, Leu52, Val80 and the methyl group of Thr44 of the receptor. Replacing reduces affinity by three orders of magnitude (the second most important residue for binding). This is surprising, as it is . One reason for its importance might be a .
Most of the residues involved in the IFNa2-IFNAR2 interaction are also found in the IFNw-IFNAR2 interface of the IFNw ternary complex.
A significant difference in the IFNAR2 interface between and IFNw is related to , which is replaced with Lys152 in . In the , this residue forms an with Glu149(IFN), but .
IFNAR1-IFN interaction
Because of the lower resolution of the IFNa ternary complex, we focused on the in our analysis of the IFN-IFNAR1 interface. In the , the only contains two hotspot residues we could experimentally confirm, . Substituting these residues by alanine reduces the affinity to all tested IFN ligands by more than 10-fold. On , mutation studies have shown that a charge-reversal (Arg 120 on IFNa) leads to a total loss of activity.
Indeed, this residue forms a in addition to . Substitution of glutamate for Arg123(IFN) would lead to electrostatic repulsion with Asp132(IFNAR1).
The low affinity of IFNAR1 for the ligand appears to be functionally relevant, as weak binding to IFNAR1 is conserved between all alpha IFNs. Three amino acid substitutions on IFNa2 at positions His57, Glu58 and Ser61 to alanine or to Tyr, Asn, and Ser, respectively, confer tighter binding to IFNAR1, but leave the affinity to IFNAR2 essentially unaltered.
Implications for the binding mode of IFNb
30% and 33% sequence identity with and IFNa2, respectively. in our ternary complex structure leads of side chains ( and ) with the receptors, indicating that the IFNb ligand could be easily accommodated by the receptors in a position similar to IFNw and IFNa2. Furthermore, the shows that . As a result, Ala19(IFN), when mutated to tryptophan, promotes an increased binding affinity to IFNAR2, which is a result of the (as shown by double mutant cycle analysis).
Structural Movements
Structural pertubations upon binding
One of the more controversial aspects of cytokine signaling is whether receptor binding is sufficient to generate activity, or if it has to be accompanied by structural perturbations. The type I interferon signaling complex is a rare example of a cytokine receptor complex were the structures of all the components making up the biologically active complex were determined to high resolution in both their free and bound forms. of the unbound NMR structure with the ternary complex structure of interferon shows a small expansion during complex formation.
Both IFNAR1 and IFNAR2, however, undergo significant domain movements upon binding. Using the D1 domain as anchor, a of IFNAR2 upon binding, on a scale of 6-12 Å, is observed (comparison of the unbound receptor (1n6u) with the binary IFNa2-IFNAR2 complex). The superimposition of the IFNa2-IFNAR2 binary complex with IFN-IFNAR2 in the ternary complexes of 6-9 Å, and even between the ternary IFNa and IFNw complexes a movement of 3-5 Å is observed. The D2 domain is engaged in crystal contacts in all three structures, and it remains an open question if the conformational changes in IFNAR2 are physiologically relevant. Still, these movements could change the proximity or orientation of the ICDs and associated Jaks within the cell.
The low-affinity receptor chain, IFNAR1, also upon ligand binding. When using D1 as anchor, D3 is moving inwards (toward the ligand) by ~15 Å. This would generate an even larger movement of the membrane-proximal SD4 domain and the transmembrane helix. The conformational changes in IFNAR1 are necessary to form the full spectrum of interactions with the IFN ligand, and to form a stable signaling complex that is able to instigate downstream signaling. In contrast to SD3, SD4 seems to be highly flexible (even more than D2 of IFNAR2). One might suggest that the conformational changes in IFNAR1 by itself will be responsible for a reduced binding affinity of IFNAR1 and may slow down the rate of ligand association to IFNAR1 directly from solution.