We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Tom Sandbox
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
===GCN4 - Leucine Zipper=== | ===GCN4 - Leucine Zipper=== | ||
| - | + | Blah blah information about leucine zipper GCN4. | |
{{STRUCTURE_1ysa| PDB=1ysa | SCENE= }} | {{STRUCTURE_1ysa| PDB=1ysa | SCENE= }} | ||
==About this Structure== | ==About this Structure== | ||
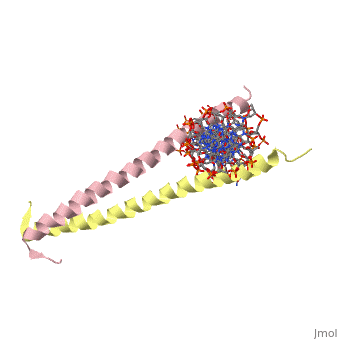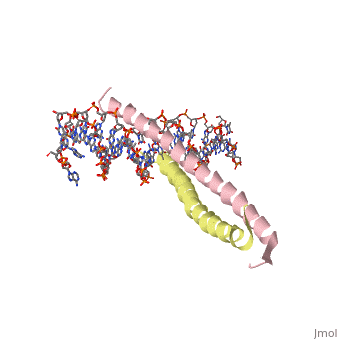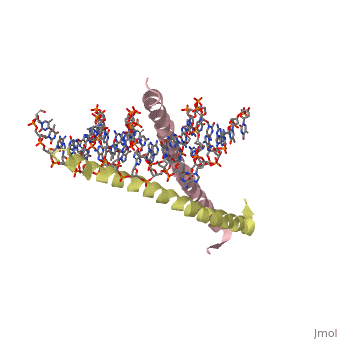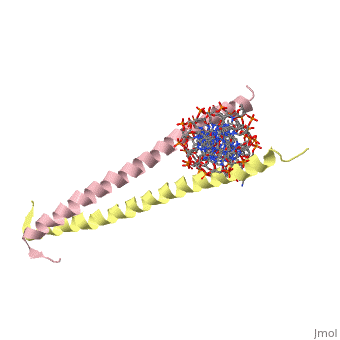
| - | [[2zta]] is a | + | [[2zta]] is composed of two identical 52 residue alpha helix chains that grouped together to form a dimer. The dimer binds through interlocking leucine amino acids in the C terminal ends, while pinching in on the major groove of DNA in the N terminal end. |
==See Also== | ==See Also== | ||
| - | + | [[2zta]] | |
| + | [[1ysa]] | ||
==Reference== | ==Reference== | ||
| - | <ref group="xtra">PMID:1557122</ref><references group="xtra"/> | ||
| - | [[Category: Saccharomyces cerevisiae]] | ||
| - | [[Category: Carey, M.]] | ||
| - | [[Category: Harrison, S C.]] | ||
| - | [[Category: Marmorstein, R.]] | ||
| - | [[Category: Ptashne, M.]] | ||
| - | [[Category: Double helix]] | ||
| - | [[Category: Protein-dna complex]] | ||
| - | [[Category: Transcription/dna complex]] | ||
Revision as of 00:34, 9 November 2011
| |||||||
| 2zta, resolution 1.80Å () | |||||||
|---|---|---|---|---|---|---|---|
| Non-Standard Residues: | |||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
Contents |
GCN4 - Leucine Zipper
Blah blah information about leucine zipper GCN4.
| |||||||||
| 1ysa, resolution 2.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
About this Structure
2zta is composed of two identical 52 residue alpha helix chains that grouped together to form a dimer. The dimer binds through interlocking leucine amino acids in the C terminal ends, while pinching in on the major groove of DNA in the N terminal end.