Tom Sandbox
From Proteopedia
| Line 8: | Line 8: | ||
| - | + | GCN4 (PDB [[2zta]] by itself, [[1ysa]] bound to DNA) is a eukaryotic transcription factor first isolated from Saccharomyces cerevisiae, also known as Baker's Yeast. | |
| + | |||
{{STRUCTURE_1ysa| PDB=1ysa | SCENE= }} | {{STRUCTURE_1ysa| PDB=1ysa | SCENE= }} | ||
| Line 14: | Line 15: | ||
===Structure=== | ===Structure=== | ||
| - | GCN4 | + | GCN4 is composed of two identical 58 residue alpha helix chains that grouped together to form a parallel coiled-coil dimer. The dimer binds through interlocking leucine amino acids and hydrophobic residues near the C terminus, while pinching in on the major groove of DNA in the N terminal end via basic residues. The basic residues are the reason the class of binding interactions is commonly referred to as bZIP or basic region leucine zipper proteins<ref name="Voet"> Voet, Donald; Voet, Judith G.; Pratt, Charlotte W. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd Ed. Hoboken, NJ: Wiley, 2008. </ref>. The X-ray structure of the 33-residue polypeptide corresponding to the leucine zipper of GCN4 was determined by Peter Kim and Thomas Alber. |
====Heptad Repeat==== | ====Heptad Repeat==== | ||
| Line 22: | Line 23: | ||
| - | '''Image 1:A pictorial representation of the heptad repeat. By following through the wheels alphabetically, it is clear how the leucines will stack every 7 units. The apostrophe denotes the | + | '''Image 1:A pictorial representation of the heptad repeat between the two subunits in a coiled-coil conformation. By following through the wheels alphabetically, it is clear how the leucines will stack every 7 units. The apostrophe denotes the diference between the two polypeptide subunits.'''(from Kgutwin in Wikipedia Commons http://commons.wikimedia.org/wiki/File:Coiledcoil-wheelcartoon.gif) |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | ===Binding=== | ||
| + | At every d and d' we have a Leucine, while at the a and a' locations we typically see valine. <ref name="Voet" /> In the opposite positions one tends to see more charged and polar residues. The Leucine and Valine repeats create a strongly hydrophobic region between the two alpha helices. | ||
====The Leucine Zipper==== | ====The Leucine Zipper==== | ||
| - | + | ===Binding with DNA=== | |
Revision as of 03:12, 9 November 2011
| |||||||
| 2zta, resolution 1.80Å () | |||||||
|---|---|---|---|---|---|---|---|
| Non-Standard Residues: | |||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
Contents |
GCN4 - The Leucine Zipper
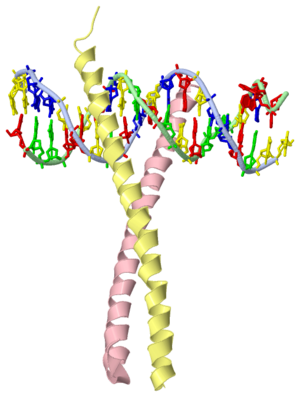
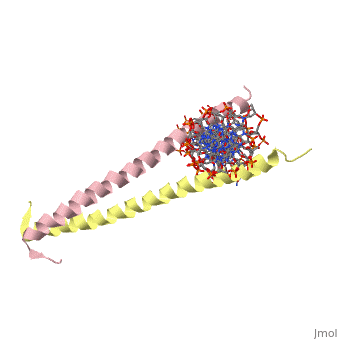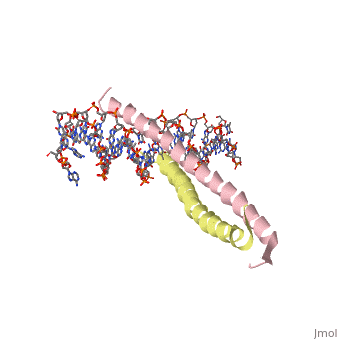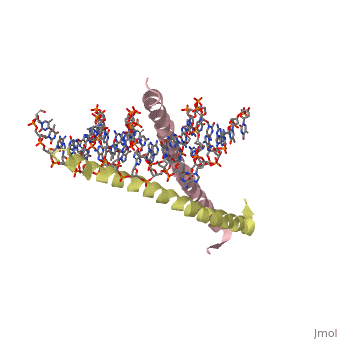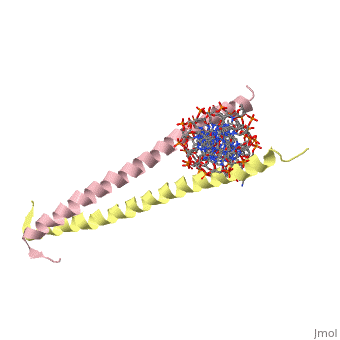
GCN4 (PDB 2zta by itself, 1ysa bound to DNA) is a eukaryotic transcription factor first isolated from Saccharomyces cerevisiae, also known as Baker's Yeast.
| |||||||||
| 1ysa, resolution 2.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Structure
GCN4 is composed of two identical 58 residue alpha helix chains that grouped together to form a parallel coiled-coil dimer. The dimer binds through interlocking leucine amino acids and hydrophobic residues near the C terminus, while pinching in on the major groove of DNA in the N terminal end via basic residues. The basic residues are the reason the class of binding interactions is commonly referred to as bZIP or basic region leucine zipper proteins[1]. The X-ray structure of the 33-residue polypeptide corresponding to the leucine zipper of GCN4 was determined by Peter Kim and Thomas Alber.
Heptad Repeat
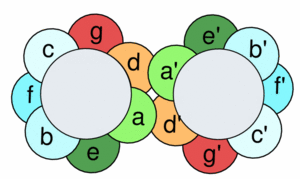
GCN4 is said to have a heptad repeat, or seven unit repeated sequence. Here the heptad repeat is a leucine, every seven units. When the structure of the alpha helices is taken into account, it is clear that each leucine is being stacked on top of the last one, every seven units. This creates the zipper-like repeated projections from the two polypeptide chains. Below we see a pictorial representation of the sequence of amino acids as we look down the length of the coiled coil structure.
Image 1:A pictorial representation of the heptad repeat between the two subunits in a coiled-coil conformation. By following through the wheels alphabetically, it is clear how the leucines will stack every 7 units. The apostrophe denotes the diference between the two polypeptide subunits.(from Kgutwin in Wikipedia Commons http://commons.wikimedia.org/wiki/File:Coiledcoil-wheelcartoon.gif)
At every d and d' we have a Leucine, while at the a and a' locations we typically see valine. [1] In the opposite positions one tends to see more charged and polar residues. The Leucine and Valine repeats create a strongly hydrophobic region between the two alpha helices.