Sandbox 39
From Proteopedia
| Line 19: | Line 19: | ||
In addition, the entirety of the secondary structure of papain can be traced from the <scene name='Sandbox_39/N_to_c_rainbow/1'>N- to the C-terminus.</scene> The red end begins the protein at the | In addition, the entirety of the secondary structure of papain can be traced from the <scene name='Sandbox_39/N_to_c_rainbow/1'>N- to the C-terminus.</scene> The red end begins the protein at the | ||
N-terminus, and can be traced through the colors of the rainbow to the purple end at the C-terminus. | N-terminus, and can be traced through the colors of the rainbow to the purple end at the C-terminus. | ||
| + | |||
| + | <Structure load='9pap' size='300' frame='true' align='left' caption='Ribbon Structure of Papain' scene=/> | ||
== Secondary Structure == | == Secondary Structure == | ||
Revision as of 04:31, 9 November 2011
| Please do NOT make changes to this Sandbox. Sandboxes 30-60 are reserved for use by Biochemistry 410 & 412 at Messiah College taught by Dr. Hannah Tims during Fall 2012 and Spring 2013. |
|
Contents |
Papain
Introduction
Papain (9PAP), also known as papaya proteinase I, is an enzyme found in unripe papaya fruit. A cysteine protease, it has been used to break down tough muscle fibers, and hence is often found in powdered meat tenderizers. It is collected from the fruit by scoring its skin and allowing the "sap" to seem out. The sap is then dried and purified.
Function
As a cysteine protease, Papain utilizes a nucleophilic cysteine thiol as part of its catalytic triad. Papain's Cys-25 is deprotonated by its His-159. The now nucleophilic Cys-25 attacks the carbonyl carbon of the peptide backbone, forming an acyl enzyme intermediate in which the peptide's amino terminal is free. Also in this step, His-159 is returned to its deprotonated form. The intermediate is then deacylated by a water molecule, and it releases the carboxyl terminal of the peptide to produce the product and regenerate the active enzyme. This entire mechanism is shown below:
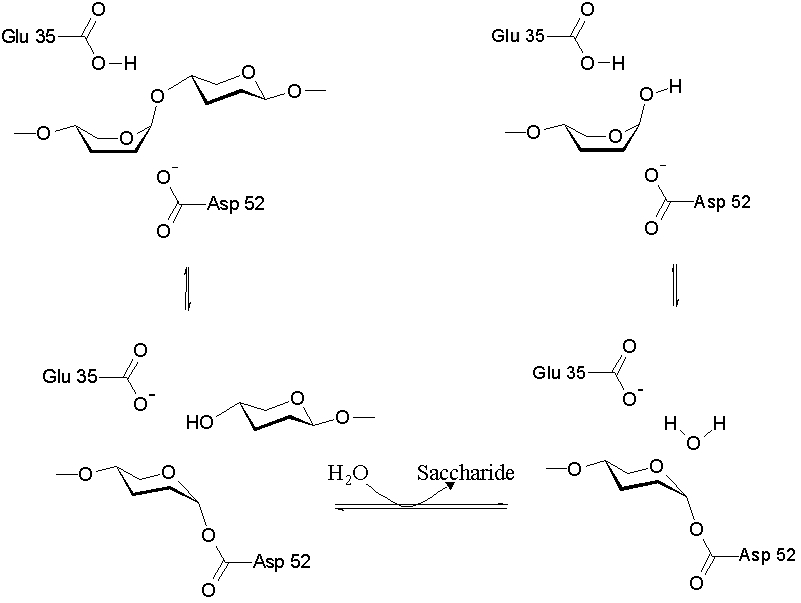 [1]]]
[1]]]
Composition of Papain
Proteins only consist of certain elements: carbon, hydrogen, nitrogen, oxygen, and sulfur. Enzymes' primary structures allow them to fold optimally and interact with their substrates maximally in order to efficiently catalyze biological reactions. The shows carbon atoms outlined in grey, oxygen atoms in red, nitrogen atoms in blue, and sulfur atoms in yellow. In addition, the entirety of the secondary structure of papain can be traced from the The red end begins the protein at the N-terminus, and can be traced through the colors of the rainbow to the purple end at the C-terminus.
|
Secondary Structure
Papain's primary structure causes it to fold into different motifs that make up its secondary structure. These motifs include , shown in blue, and , shown in green. All other motifs are nonrandom, structural units, mostly simply turns. As shown, papain has 7 alpha helices and 8 beta pleated sheets.
Polarity and Hydrophobicity
Papain contains , or "water-hating" regions, and hydrophillic, or "water-loving" regions. The hydrophobic effect, or the tendency of nonpolar substances to aggregate in aqueous solution and exclude water molecules, allows proteins to fold the way they do, exposing hydrophilic residues on their outer surface while sequestering hydrophobic residues in their center. As shown, all of the are found in the center of the folded protein.
