This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox 38
From Proteopedia
(→'''Papain''') |
|||
| Line 1: | Line 1: | ||
| + | <!-- PLEASE DO NOT DELETE THIS TEMPLATE --> | ||
| + | {{Template:Oberholser_Sandbox_Reservation}} | ||
| + | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| + | |||
<!-- PLEASE DO NOT DELETE THIS TEMPLATE --> | <!-- PLEASE DO NOT DELETE THIS TEMPLATE --> | ||
{{Template:Oberholser_Sandbox_Reservation}} | {{Template:Oberholser_Sandbox_Reservation}} | ||
| Line 5: | Line 9: | ||
= '''Papain''' = | = '''Papain''' = | ||
| - | <StructureSection load='9PAP' size='500' side='right' caption= | ||
| - | == | + | ==Introduction== |
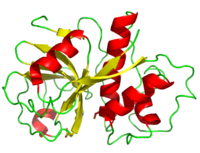
| + | [[Image:Papain_cartoon.png|200px|left|thumb|Cartoon Peak at Pepsin]] | ||
| - | + | DID YOU KNOW? | |
| - | + | <scene name='Sandbox_35/Papain/1'>Papain</scene>. Meat tenderizer. Old time home remedy for insect, jellyfish, and stingray stings<ref>[http://www.ameriden.com/products/advanced-digestive-enzyme/] Ameridan International</ref>. Who would have thought that a sulfhydryl protease from the latex of the papaya fruit, ''Carica papaya'' and ''Vasconcellea cundinamarcensis'' would have such a practical application beyond proteopedia? | |
| - | Papain was originally discovered during the colonial period in Congo. The native inhabitants discovered that wrapping their elephant meat in papaya leaves helped to tenderize the meat. While they did not know the direct cause, this was when the proteolytic enzyme was first discovered. The active binding site of this enzyme was first discovered by Drenth et al., through the crystallographic analysis of the enzymes structure. | ||
| - | == '''Composition of Papain:''' == | ||
| - | + | This protease belongs to an extended family of aminopeptidases, dipeptidyl peptidases, endopeptidases, and other enzymes having both exo- and endo-peptidase activity. The inactivated zymogen with N-terminal propeptide regions - helping to stabilize against alkalinity and enabling proper folding - is activated through removal of the propeptide regions <ref>PMID: 7845226</ref><ref>PMID: 12188906</ref>. | |
| - | == ''' | + | <StructureSection load='9pap' size='500' side='right' caption='Structure of Papain (PDB entry [[9PAP]])' scene=''> |
| + | ==Structure== | ||
| + | Its polypeptide chain consists of 212 amino acid residues folded to form a groove between its two domains containing the active site. | ||
| + | <scene name='Sandbox_35/Secondary_structure_papain/2'>secondary structure</scene> | ||
| - | + | <scene name='Sandbox_35/2nd_struc_papain_beta/2'>beta sheets</scene> | |
| - | <scene name=' | + | |
| + | <scene name='Sandbox_35/2nd_struc_papain_helix/2'>alpha helix</scene> <ref name="9PAP PDB">[http://www.pdb.org/pdb/explore/explore.do?structureId=9PAP]9PAP PDB</ref> | ||
| + | |||
| + | <scene name='Sandbox_35/Active_site_papain/3'>active site</scene> <ref>PMID: 8140097</ref> | ||
| + | <scene name='Sandbox_35/Active_site_asp_158_papain/1'>TextToBeDisplayed</scene> | ||
| + | |||
| + | ===Distribution of Residues=== | ||
| + | <scene name='Sandbox_35/Papain_acid_and_basic_residues/1'>acidic and basic residues</scene> | ||
| + | |||
| + | <scene name='Sandbox_35/Hydrophobicity_papain/3'>polar and non-polar residue</scene> | ||
| + | |||
| + | <scene name='Sandbox_35/Papain_polar/1'>polar residues</scene> | ||
| + | |||
| + | <scene name='Sandbox_35/Nonpolar_papain/2'>non-polar residues</scene> | ||
| + | |||
| + | ===Ligands interactions=== | ||
| + | <scene name='Sandbox_35/Cathepsin_l_specific_inhibitor/2'>Cathepsin L specific inhibitor</scene> | ||
| + | Primarily hydrogen bonds with non-water and hydrophobic interactions | ||
| + | |||
| + | <scene name='Sandbox_35/Cathepsin_interaction/3'>interaction</scene> | ||
</StructureSection> | </StructureSection> | ||
| + | |||
| + | ==Catalytic Mechanism== | ||
| + | [[Image:Papainmech6.jpg|200px|left|thumb| General mechanism of papain catalysis<ref>[http://chemistry.umeche.maine.edu/CHY431/Peptidase10.html] University of Maine</ref>.]] | ||
| + | |||
| + | ==References== | ||
| + | <references /> | ||
| + | <ref group="xtra">PMID:8140097</ref> | ||
| + | |||
| + | http://www.pdb.org/pdb/explore/explore.do?structureId=2PAD | ||
| + | • Show the secondary structures. | ||
| + | • Compare the distribution of polar residues to that of nonpolar residues. | ||
| + | • Highlight the active site. | ||
| + | • If you can find a PDB file of the enzyme that contains a pseudo-substrate (may be inhibitor), highlight it. | ||
| + | • Show the contacts or attractions that are present between the pseudo-substrate and the protein, and if the enzyme has multiple subunits, show the contacts between the subunits. | ||
| + | • Identify any other ligands that are present in the structure and the types of contacts that are present between them and the protein | ||
| + | |||
| + | http://proteopedia.org/wiki/index.php/Sandbox_55#cite_note-18 | ||
| + | Table of contents | ||
| + | Pictures | ||
| + | References (cross links) | ||
Revision as of 20:33, 13 November 2011
| Please do NOT make changes to this Sandbox. Sandboxes 30-60 are reserved for use by Biochemistry 410 & 412 at Messiah College taught by Dr. Hannah Tims during Fall 2012 and Spring 2013. |
| Please do NOT make changes to this Sandbox. Sandboxes 30-60 are reserved for use by Biochemistry 410 & 412 at Messiah College taught by Dr. Hannah Tims during Fall 2012 and Spring 2013. |
Contents |
Papain
Introduction
DID YOU KNOW?
. Meat tenderizer. Old time home remedy for insect, jellyfish, and stingray stings[1]. Who would have thought that a sulfhydryl protease from the latex of the papaya fruit, Carica papaya and Vasconcellea cundinamarcensis would have such a practical application beyond proteopedia?
This protease belongs to an extended family of aminopeptidases, dipeptidyl peptidases, endopeptidases, and other enzymes having both exo- and endo-peptidase activity. The inactivated zymogen with N-terminal propeptide regions - helping to stabilize against alkalinity and enabling proper folding - is activated through removal of the propeptide regions [2][3].
| |||||||||||
Catalytic Mechanism
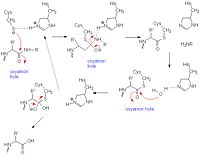
References
- ↑ [1] Ameridan International
- ↑ Rawlings ND, Barrett AJ. Families of cysteine peptidases. Methods Enzymol. 1994;244:461-86. PMID:7845226
- ↑ Yamamoto Y, Kurata M, Watabe S, Murakami R, Takahashi SY. Novel cysteine proteinase inhibitors homologous to the proregions of cysteine proteinases. Curr Protein Pept Sci. 2002 Apr;3(2):231-8. PMID:12188906
- ↑ [2]9PAP PDB
- ↑ Wang J, Xiang YF, Lim C. The double catalytic triad, Cys25-His159-Asp158 and Cys25-His159-Asn175, in papain catalysis: role of Asp158 and Asn175. Protein Eng. 1994 Jan;7(1):75-82. PMID:8140097
- ↑ [3] University of Maine
http://www.pdb.org/pdb/explore/explore.do?structureId=2PAD
• Show the secondary structures.
• Compare the distribution of polar residues to that of nonpolar residues.
• Highlight the active site.
• If you can find a PDB file of the enzyme that contains a pseudo-substrate (may be inhibitor), highlight it.
• Show the contacts or attractions that are present between the pseudo-substrate and the protein, and if the enzyme has multiple subunits, show the contacts between the subunits.
• Identify any other ligands that are present in the structure and the types of contacts that are present between them and the protein
http://proteopedia.org/wiki/index.php/Sandbox_55#cite_note-18 Table of contents Pictures References (cross links)