User:Mitchell Long/Sandbox 1
From Proteopedia
| Line 1: | Line 1: | ||
| - | + | <StructureSection load='3fgc' size='500' side='right' caption='Structure of Bacterial Luciferase and FMN complex from ''V. harveyi'' (PDB entry [[3fgc]])' scene=''> | |
| + | |||
| + | ==Introduction== | ||
| + | Luciferases are a class of enzymes that catalyze the oxidation of a long chain aliphatic aldehydes. This reaction results in the formation of a carboxylic acid and the emission of photons in the form of blue-green light. | ||
| + | |||
| + | |||
| + | |||
| + | |||
</StructureSection> | </StructureSection> | ||
| - | + | ||
{{STRUCTURE_3fgc| PDB=3fgc | SCENE= }} | {{STRUCTURE_3fgc| PDB=3fgc | SCENE= }} | ||
| - | ==Mechanism of Bio luminescence == | + | ==Mechanism of Bio luminescence== |
| + | Luciferase found in ''''Italic tex<StructureSection load='3fgc' size='500' side='right' caption='Structure of Bacterial Luciferase and FMN complex from ''V. harveyi'' (PDB entry [[3fgc]])' scene=''> | ||
| + | |||
| + | ==Introduction== | ||
| + | Luciferases are a class of enzymes that catalyze the oxidation of a long chain aliphatic aldehydes. This reaction results in the formation of a carboxylic acid and the emission of photons in the form of blue-green light. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </StructureSection> | ||
| + | |||
| + | |||
| + | {{STRUCTURE_3fgc| PDB=3fgc | SCENE= }} | ||
| + | |||
| + | |||
| + | |||
| + | ==Mechanism of Bio luminescence== | ||
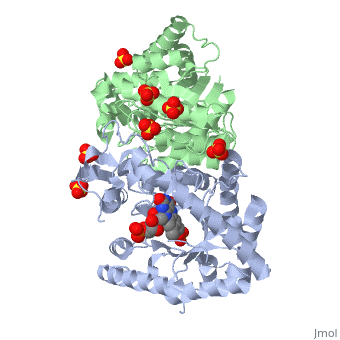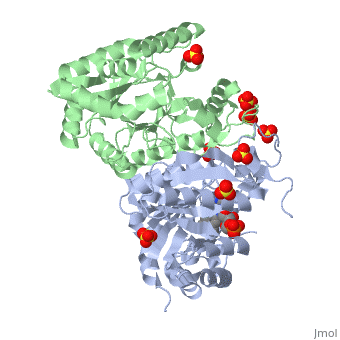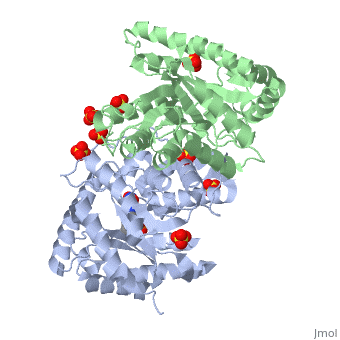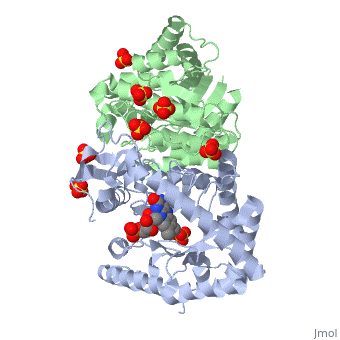
| + | Luciferase found in ''''Vibrio harveyi'''' is a heterodimeric flavin monoxygenase that is composed of a catalytic α subunit and a homologous but noncatalytic β subunit. The catalytic α subunit is bound to a Flavin Mononucleotide cofactor. Bioluminescense is achieved by a two step reaction between | ||
| + | |||
| + | |||
==Structural Motifs== | ==Structural Motifs== | ||
| + | Active Site | ||
| + | |||
| + | Mobile Loop-The mobile loop of luciferase is thought to be responsible for protecting reaction intermediates and preventing the entry of bulk solvent into the active center once the flavin substrate is bound. | ||
| + | |||
| + | (β/α)&sub8; Barrel- | ||
| + | |||
| + | Interface between Alpha and Beta Subunits | ||
| + | |||
| + | |||
| + | |||
==Applications In Biotechnology== | ==Applications In Biotechnology== | ||
| + | The luciferase assay | ||
==Quorum Sensing== | ==Quorum Sensing== | ||
| + | Quorum sensing is a population density dependent form gene expression found in many bacteria. | ||
Revision as of 03:12, 17 November 2011
| |||||||||||
| |||||||||
| 3fgc, resolution 2.30Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | luxA (Vibrio harveyi), luxB (Vibrio harveyi) | ||||||||
| Activity: | Alkanal monooxygenase (FMN-linked), with EC number 1.14.14.3 | ||||||||
| Related: | 1luc, 1brl | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Mechanism of Bio luminescence
Luciferase found in 'Italic tex
| |||||||||||
| |||||||||
| 3fgc, resolution 2.30Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | luxA (Vibrio harveyi), luxB (Vibrio harveyi) | ||||||||
| Activity: | Alkanal monooxygenase (FMN-linked), with EC number 1.14.14.3 | ||||||||
| Related: | 1luc, 1brl | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Mechanism of Bio luminescence
Luciferase found in 'Vibrio harveyi' is a heterodimeric flavin monoxygenase that is composed of a catalytic α subunit and a homologous but noncatalytic β subunit. The catalytic α subunit is bound to a Flavin Mononucleotide cofactor. Bioluminescense is achieved by a two step reaction between
Structural Motifs
Active Site
Mobile Loop-The mobile loop of luciferase is thought to be responsible for protecting reaction intermediates and preventing the entry of bulk solvent into the active center once the flavin substrate is bound.
(β/α)&sub8; Barrel-
Interface between Alpha and Beta Subunits
Applications In Biotechnology
The luciferase assay
Quorum Sensing
Quorum sensing is a population density dependent form gene expression found in many bacteria.