This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
A-ATP Synthase
From Proteopedia
| Line 35: | Line 35: | ||
<scene name='A-ATP_Synthase/Vanadate_1_interactions/2'>Vanadate one</scene> occupies the ADP site. Although not at bonding distances the residues P233 G234 L417 stabilize the first vanadate in the transition state with weak nonpolar interactions. Residues K240 and T241 stabilize with polar interactions. | <scene name='A-ATP_Synthase/Vanadate_1_interactions/2'>Vanadate one</scene> occupies the ADP site. Although not at bonding distances the residues P233 G234 L417 stabilize the first vanadate in the transition state with weak nonpolar interactions. Residues K240 and T241 stabilize with polar interactions. | ||
| - | Residue <scene name='A-ATP_Synthase/238/ | + | Residue <scene name='A-ATP_Synthase/238/6'>S238</scene> is a polar serine molecule that interacts with the nucleotides via a hydrogen bond during catalysis. The distance between residue S238 is longest in '''As''', shortest in '''Avi''' and intermediate in '''Apnp''' . In '''As''' a water molecule bridges the gap, which is removed in '''Avi'''. Dehydration of the transition state active site is reversed when ATP forms. In '''Apnp''' the water molecule interacts with the y-phosphate of ATP. |
==Comparasons to other known structures== | ==Comparasons to other known structures== | ||
Revision as of 23:55, 29 November 2011
Structure
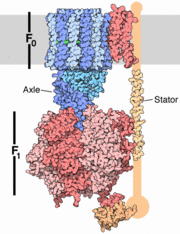
The structure on the right shows the F1 motor and the axle that connects the two. ATP synthesis is composed of two rotary motors, each powered by a different fuel. The motor at the top, termed F0, an electric motor. It is embedded in a membrane (shown schematically as a gray stripe here), and is powered by the flow of hydrogen ions across the membrane. As the protons flow through the motor, they turn a circular rotor . This rotor is connected to the second motor, termed F1. The F1 motor is a chemical motor, powered by ATP. The two motors are connected together by a stator, shown on the right, so that when F0 turns, F1 turns too. A-ATP synthase is very similar to F ATP Synthase and is composed of two parts A1 and A0 which are composed of at least nine subunits A3B3C:D:E:F:H2:a:cx that function as a pair of rotary motors connected by central and peripheral stalk(s) [1]. The A0 domain is the hydrophobic membrane embedded ion-translocating sector that uses the H+ gradient to power ATP synthase in domain A1. A1 is catalytic and water soluble containing A and B subunits. These subunits are comparable to F-ATP synthase ATP synthase alpha/beta subunits. The A subunit of A1 is catalytic and the B subunit is regulatory, with a substrate-binding site on each. [1]
| |||||||||||
| |||||||||||
References
- ↑ 1.0 1.1 Muller V, Lemker T, Lingl A, Weidner C, Coskun U, Gruber G. Bioenergetics of archaea: ATP synthesis under harsh environmental conditions. J Mol Microbiol Biotechnol. 2005;10(2-4):167-80. PMID:16645313 doi:10.1159/000091563
- ↑ Gibbons C, Montgomery MG, Leslie AG, Walker JE. The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution. Nat Struct Biol. 2000 Nov;7(11):1055-61. PMID:11062563 doi:10.1038/80981
- ↑ Schafer IB, Bailer SM, Duser MG, Borsch M, Bernal RA, Stock D, Gruber G. Crystal structure of the archaeal A1Ao ATP synthase subunit B from Methanosarcina mazei Go1: Implications of nucleotide-binding differences in the major A1Ao subunits A and B. J Mol Biol. 2006 May 5;358(3):725-40. Epub 2006 Mar 10. PMID:16563431 doi:http://dx.doi.org/10.1016/j.jmb.2006.02.057
- ↑ Gonzalez JM, Masuchi Y, Robb FT, Ammerman JW, Maeder DL, Yanagibayashi M, Tamaoka J, Kato C. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles. 1998 May;2(2):123-30. PMID:9672687
- ↑ 5.0 5.1 Manimekalai MS, Kumar A, Jeyakanthan J, Gruber G. The Transition-Like State and P(i) Entrance into the Catalytic A Subunit of the Biological Engine A-ATP Synthase. J Mol Biol. 2011 Mar 16. PMID:21396943 doi:10.1016/j.jmb.2011.03.010
- ↑ Priya R, Kumar A, Manimekalai MS, Gruber G. Conserved Glycine Residues in the P-Loop of ATP Synthases Form a Doorframe for Nucleotide Entrance. J Mol Biol. 2011 Sep 8. PMID:21925186 doi:10.1016/j.jmb.2011.08.045
Proteopedia Page Contributors and Editors (what is this?)
Kaitlin Chase MacCulloch, Michal Harel, Alexander Berchansky