Sandbox Reserved 383
From Proteopedia
(Replacing page with '<!-- PLEASE DO NOT DELETE THIS TEMPLATE --> {{Sandbox_Reserved_JMeans}} <!-- PLEASE ADD YOUR CONTENT BELOW HERE -->') |
|||
| Line 2: | Line 2: | ||
{{Sandbox_Reserved_JMeans}} | {{Sandbox_Reserved_JMeans}} | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
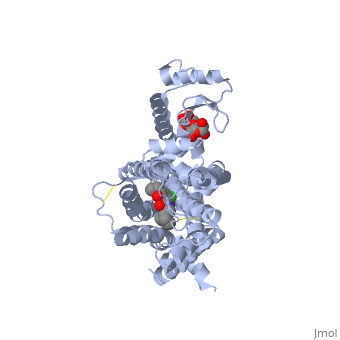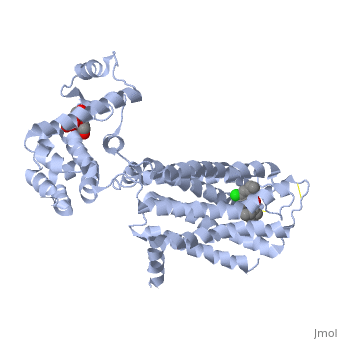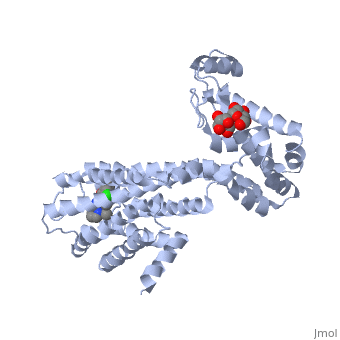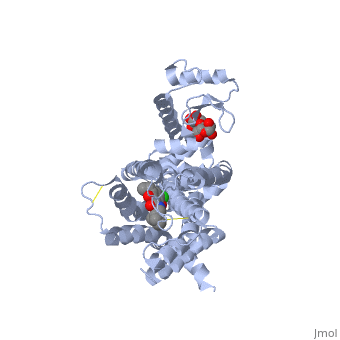
| + | <Structure load='3PBL' size='350' frame='true' align='right' caption='Human dopamine D3 receptor in complex with eticlopride' scene='Human dopamine D3 receptor in complex with eticlopride/> | ||
| + | ==Background== | ||
| + | Human dopamine D3 receptor is a protein that encoded by the dopamine receptor gene (DRD3).<ref>Le Coniat M, Sokoloff P, Hillion J, Martres MP, Giros B, Pilon C, Schwartz JC, Berger R (Oct 1991).</ref> The DRD3 gene codes for the D3 dopamine receptor that inhibits adenylyl cyclase through inhibitory G-proteins. This receptor is located in the brain, suggesting that the D3 receptor plays a role in cognitive and emotional functions.<ref>http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=1814.</ref> The human dopamine D3 receptor is membrane-bound and scattered in the cytoplasm. Receptor stimulation causes internalization of the receptors at the perinuclear ares. This is followed by the spreading of the receptors to the membrane. DRD3 is contained in lipid rafts of renal proximal tubule cells.<ref> http://www.uniprot.org/uniprot/P35462</ref> The crystal structure of the human dopamine D3 receptor (D3R) in complex with the small molecule D2R/D3R-specific antagonist eticlopride reveals important features of the ligand binding pocket and extracellular loops. On the intracellular side of the receptor, a locked conformation of the ionic lock and two distinctly different conformations of intracellular loop 2 are observed. Docking of R-22, a D3R-selective antagonist, reveals an extracellular extension of the eticlopride binding site that comprises a second binding pocket for the aryl amide of R-22, which differs between the highly homologous D2R and D3R. This difference provides direction to the design of D3R-selective agents for treating drug abuse and other neuropsychiatric indications.<ref>http://www.pdb.org/pdb/explore.do?structureId=3PBL</ref> | ||
| + | ==Ligands== | ||
| + | Ligands that are associated with dopamine D3 receptor are 3-chloro-5-ethyl-N{[(2S)-1-ethylpyrrolidin-2-yl]methyl}-6-hydroxyl-2-methyloxybenzamide (ETQ on PDB) and maltose (MAL on PDB). ETQ binds to dopamine D3 receptor by Asp 110A and Phe346A. MAL binds by Asp 1020A, Glu 1022A, Glu1011A, and Leu1032A.<ref>http://www.pdb.org/pdb/explore.do?structureId=3PBL</ref> | ||
| + | ==Disease Involvement== | ||
| + | Variations in the DRD3 gene is connected with essential tremor hereditary type 1 (ETM1). ETM1 is the most common movement disorder involving postural tremor of the arms, head, legs, body core, voice, jaw, and other facial muscles. This condition can be provoked by emotions, hunger, fatigue, and temperature extremes.<ref>http://www.uniprot.org/uniprot/P35462</ref> Disorders that are linked to variations in the DRD3 gene include social phobia, Tourette’s syndrome, Parkinson’s disease, schizophrenia, neuroleptic malignant syndrome, attention-deficit hyperactivinty disorder (ADHD), and drug and alcohol dependence.<ref>http://en.wikipedia.org/wiki/Dopamine_receptor</ref> | ||
| + | ==References== | ||
| + | # <Le Coniat M, Sokoloff P, Hillion J, Martres MP, Giros B, Pilon C, Schwartz JC, Berger R (Oct 1991). "Chromosomal localization of the human D3 dopamine receptor gene". Hum Genet 87 (5): 618–20./> | ||
| + | # <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=1814/> | ||
| + | # <http://www.uniprot.org/uniprot/P35462/> | ||
| + | # <http://www.pdb.org/pdb/explore.do?structureId=3PBL/> | ||
| + | # <http://www.pdb.org/pdb/explore.do?structureId=3PBL/> | ||
| + | # <http://www.uniprot.org/uniprot/P35462/> | ||
| + | # <http://en.wikipedia.org/wiki/Dopamine_receptor/> | ||
Revision as of 22:50, 30 November 2011
| This Sandbox is Reserved from September 14, 2021, through May 31, 2022, for use in the class Introduction to Biochemistry taught by User:John Means at the University of Rio Grande, Rio Grande, OH, USA. This reservation includes 5 reserved sandboxes (Sandbox Reserved 1590 through Sandbox Reserved 1594). |
To get started:
More help: Help:Editing. For an example of a student Proteopedia page, please see Photosystem II, Tetanospasmin, or Guanine riboswitch. |
|
Contents |
Background
Human dopamine D3 receptor is a protein that encoded by the dopamine receptor gene (DRD3).[1] The DRD3 gene codes for the D3 dopamine receptor that inhibits adenylyl cyclase through inhibitory G-proteins. This receptor is located in the brain, suggesting that the D3 receptor plays a role in cognitive and emotional functions.[2] The human dopamine D3 receptor is membrane-bound and scattered in the cytoplasm. Receptor stimulation causes internalization of the receptors at the perinuclear ares. This is followed by the spreading of the receptors to the membrane. DRD3 is contained in lipid rafts of renal proximal tubule cells.[3] The crystal structure of the human dopamine D3 receptor (D3R) in complex with the small molecule D2R/D3R-specific antagonist eticlopride reveals important features of the ligand binding pocket and extracellular loops. On the intracellular side of the receptor, a locked conformation of the ionic lock and two distinctly different conformations of intracellular loop 2 are observed. Docking of R-22, a D3R-selective antagonist, reveals an extracellular extension of the eticlopride binding site that comprises a second binding pocket for the aryl amide of R-22, which differs between the highly homologous D2R and D3R. This difference provides direction to the design of D3R-selective agents for treating drug abuse and other neuropsychiatric indications.[4]
Ligands
Ligands that are associated with dopamine D3 receptor are 3-chloro-5-ethyl-N{[(2S)-1-ethylpyrrolidin-2-yl]methyl}-6-hydroxyl-2-methyloxybenzamide (ETQ on PDB) and maltose (MAL on PDB). ETQ binds to dopamine D3 receptor by Asp 110A and Phe346A. MAL binds by Asp 1020A, Glu 1022A, Glu1011A, and Leu1032A.[5]
Disease Involvement
Variations in the DRD3 gene is connected with essential tremor hereditary type 1 (ETM1). ETM1 is the most common movement disorder involving postural tremor of the arms, head, legs, body core, voice, jaw, and other facial muscles. This condition can be provoked by emotions, hunger, fatigue, and temperature extremes.[6] Disorders that are linked to variations in the DRD3 gene include social phobia, Tourette’s syndrome, Parkinson’s disease, schizophrenia, neuroleptic malignant syndrome, attention-deficit hyperactivinty disorder (ADHD), and drug and alcohol dependence.[7]
References
- <Le Coniat M, Sokoloff P, Hillion J, Martres MP, Giros B, Pilon C, Schwartz JC, Berger R (Oct 1991). "Chromosomal localization of the human D3 dopamine receptor gene". Hum Genet 87 (5): 618–20./>
- <http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=1814/>
- <http://www.uniprot.org/uniprot/P35462/>
- <http://www.pdb.org/pdb/explore.do?structureId=3PBL/>
- <http://www.pdb.org/pdb/explore.do?structureId=3PBL/>
- <http://www.uniprot.org/uniprot/P35462/>
- <http://en.wikipedia.org/wiki/Dopamine_receptor/>