Sandbox 666
From Proteopedia
| Line 1: | Line 1: | ||
Hello this Sandbox is reserved for a student project | Hello this Sandbox is reserved for a student project | ||
| - | + | {{STRUCTURE_1eri| PDB=1eri | SCENE= | size='500'}} | |
== Presentation == | == Presentation == | ||
| - | + | ||
EcoRI is a type II restriction endonuclease. It recognizes and cleaves DNA on a specific palindromic sequence: GAATTC. EcoRI has been extracted from strain R Escherichia coli, a common bacterium which populates the intestine of mammalians. In bacteria, restriction enzymes protect the cell by cutting foreign DNA from bacteriophages (specific bacterial viruses). | EcoRI is a type II restriction endonuclease. It recognizes and cleaves DNA on a specific palindromic sequence: GAATTC. EcoRI has been extracted from strain R Escherichia coli, a common bacterium which populates the intestine of mammalians. In bacteria, restriction enzymes protect the cell by cutting foreign DNA from bacteriophages (specific bacterial viruses). | ||
| Line 9: | Line 9: | ||
| - | EcoRI (E.C. 3.1.21.4) is a hydrolase and its substrate is a double-strand DNA molecule and two water molecules. For its catalytic activity, EcoRI needs a cofactor which is the divalent ion | + | EcoRI (E.C. 3.1.21.4) is a hydrolase and its substrate is a double-strand DNA molecule and two water molecules. For its catalytic activity, EcoRI needs a cofactor which is the divalent ion Mg<sup>2+</sup>. EcoRI hydrolyses the phosphodiester bond between the guanylic and adenylic residues resulting in 5’-phosphate sticky ends which are complementary. |
== Structure == | == Structure == | ||
| - | EcoRI is a homodimer, so it has two identical subunits (Representation of one <scene name='Sandbox_666/Monomer_structure/3'>subunit</scene>) of 31 kDa, but it possible to have homotetramers at high concentrations. The constitutive monomers are 276 amino acids long. EcoRI and all the other restriction enzymes show a common structural core which is a α/β domain. The constitutive subunits of EcoRI are organized into a single α/β domain (five stranded <scene name='Sandbox_666/B_sheet/1'>β</scene> sheet which | + | EcoRI is a homodimer, so it has two identical subunits (Representation of one <scene name='Sandbox_666/Monomer_structure/3'>subunit</scene>) of 31 kDa, but it possible to have homotetramers at high concentrations. The constitutive monomers are 276 amino acids long. EcoRI and all the other restriction enzymes show a common structural core which is a α/β domain. The constitutive subunits of EcoRI are organized into a single α/β domain (five stranded <scene name='Sandbox_666/B_sheet/1'>β</scene> sheet which is surrounded by <scene name='Sandbox_666/Helix/1'>α helices</scene> ). Four of these five β strands are parallel whereas the fourth (β4) is in an anti-parallel orientation to the others.<ref>Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. |
| + | Kim YC, Grable JC, Love R, Greene PJ, Rosenberg JM.</ref> | ||
| Line 20: | Line 21: | ||
| - | The specific recognition of EcoRI of the GAATTC sequence is mediated by twelve hydrogen bonds (six bonds per subunit) originating from α helical recognition modules. Three amino acids are responsible for the recognition: | + | The specific recognition of EcoRI of the GAATTC sequence is mediated by twelve hydrogen bonds (six bonds per subunit) originating from α helical recognition modules. Three amino acids are responsible for the recognition:Arg<sup>200</sup>, Glu<sup>144</sup> and Arg<sup>145</sup>. These aminoacids are shown in red <scene name='Sandbox_666/Grey_protein/3'>here</scene> .Each residue form two hydrogen bonds with Guanine and the adjacent Adenosine residues respectively. |
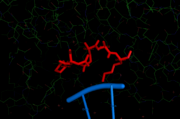
| - | The reaction is due to a catalytic sequence motif which is found in most type II restriction endonucleases: the PD…D/EXK motif. For EcoRI, this catalytic sequence is | + | The reaction is due to a catalytic sequence motif which is found in most type II restriction endonucleases: the PD…D/EXK motif. For EcoRI, this catalytic sequence is PD<sup>91</sup> …E<sup>111</sup>AK.<scene name='Sandbox_666/Catalytic_core/3'>The catalytic core is shown here in red</scene>. This motif is also responsible for Mg2+ binding(Asp90 and Glu111). [[Image:Catalytic 1ERI.png | thumb | The catalytic core of Eco RI with the target DNA]]<ref> |
| + | Structure and function of type II restriction endonucleases | ||
| + | Alfred Pingoud, Albert Jeltsch | ||
| + | Nucleic Acids Res. 2001 September 15; 29(18): 3705–3727. | ||
| + | PMCID: PMC55916</ref> | ||
| Line 31: | Line 36: | ||
Type II restriction endonuclease like EcoRI are often used in molecular biology for their capacity to cut precisely DNA on specific restriction site. That makes them useful tools for gene cloning. By using two different restriction enzymes, it is possible to do directional cloning which is very important if you want to insert a gene in an expression vector. | Type II restriction endonuclease like EcoRI are often used in molecular biology for their capacity to cut precisely DNA on specific restriction site. That makes them useful tools for gene cloning. By using two different restriction enzymes, it is possible to do directional cloning which is very important if you want to insert a gene in an expression vector. | ||
| - | + | == Links == | |
| + | [http://proteopedia.org/wiki/index.php?title=Sandbox_666&action=submit] | ||
== References == | == References == | ||
| + | <references/> | ||
| - | <rRefinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. | ||
| - | Kim YC, Grable JC, Love R, Greene PJ, Rosenberg JM. | ||
| - | Source | ||
| - | Department of Biological Sciences, University of Pittsburgh, PA 15260./> | ||
| - | |||
| - | Structure and function of type II restriction endonucleases | ||
| - | Alfred Pingoud, Albert Jeltsch | ||
| - | Nucleic Acids Res. 2001 September 15; 29(18): 3705–3727. | ||
| - | PMCID: PMC55916 | ||
<!-- | <!-- | ||
Revision as of 14:09, 16 December 2011
Hello this Sandbox is reserved for a student project
| |||||||
| 1eri, resolution 2.50Å () | |||||||
|---|---|---|---|---|---|---|---|
| |||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
Contents |
Presentation
EcoRI is a type II restriction endonuclease. It recognizes and cleaves DNA on a specific palindromic sequence: GAATTC. EcoRI has been extracted from strain R Escherichia coli, a common bacterium which populates the intestine of mammalians. In bacteria, restriction enzymes protect the cell by cutting foreign DNA from bacteriophages (specific bacterial viruses).
Reaction
EcoRI (E.C. 3.1.21.4) is a hydrolase and its substrate is a double-strand DNA molecule and two water molecules. For its catalytic activity, EcoRI needs a cofactor which is the divalent ion Mg2+. EcoRI hydrolyses the phosphodiester bond between the guanylic and adenylic residues resulting in 5’-phosphate sticky ends which are complementary.
Structure
EcoRI is a homodimer, so it has two identical subunits (Representation of one ) of 31 kDa, but it possible to have homotetramers at high concentrations. The constitutive monomers are 276 amino acids long. EcoRI and all the other restriction enzymes show a common structural core which is a α/β domain. The constitutive subunits of EcoRI are organized into a single α/β domain (five stranded sheet which is surrounded by ). Four of these five β strands are parallel whereas the fourth (β4) is in an anti-parallel orientation to the others.[1]
The N-terminal section of each subunit forms, with a β-hairpin, an “arm” which wraps around the DNA molecule. (The arm brings the DNA molecule to the catalytic cleft.)
The specific recognition of EcoRI of the GAATTC sequence is mediated by twelve hydrogen bonds (six bonds per subunit) originating from α helical recognition modules. Three amino acids are responsible for the recognition:Arg200, Glu144 and Arg145. These aminoacids are shown in red .Each residue form two hydrogen bonds with Guanine and the adjacent Adenosine residues respectively.
Application of EcoRI in molecular biology
Type II restriction endonuclease like EcoRI are often used in molecular biology for their capacity to cut precisely DNA on specific restriction site. That makes them useful tools for gene cloning. By using two different restriction enzymes, it is possible to do directional cloning which is very important if you want to insert a gene in an expression vector.
Links
References
- ↑ Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Kim YC, Grable JC, Love R, Greene PJ, Rosenberg JM.
- ↑ Structure and function of type II restriction endonucleases Alfred Pingoud, Albert Jeltsch Nucleic Acids Res. 2001 September 15; 29(18): 3705–3727. PMCID: PMC55916