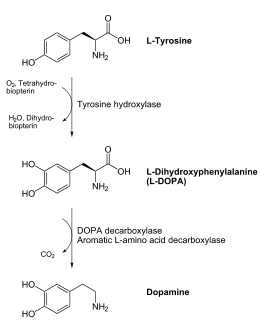
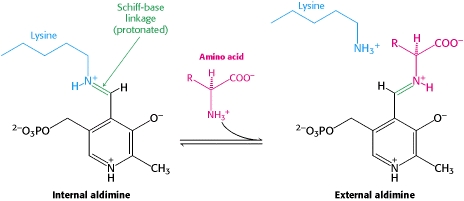
DOPA decarboxylase
From Proteopedia
(Difference between revisions)
| Line 42: | Line 42: | ||
===Inhibitor Binding=== | ===Inhibitor Binding=== | ||
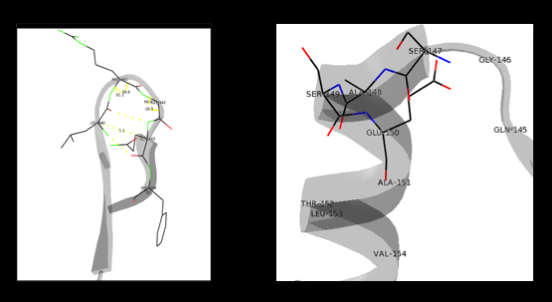
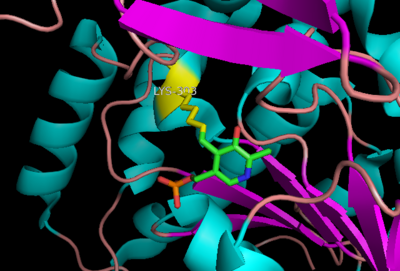
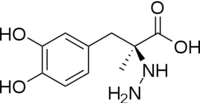
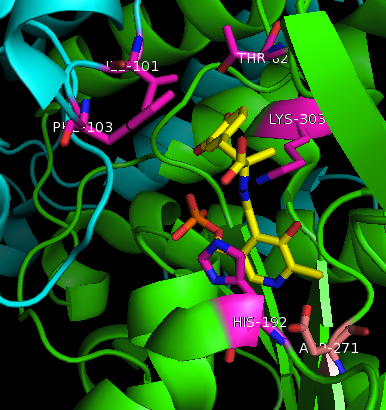
| - | [[image:actsite.png|thumb| | + | [[image:carbiDOPA.png|thumb|left|200px|'''carbiDOPA''']]The inhibitor <scene name='DOPA_decarboxylase/Carbidopa/1'>carbiDOPA</scene> binds to the enzyme by forming a hydrazone linkage with PLP through its hydrazine moiety. The catechol ring of carbiDOPA is deeply buried in the active site cleft and is stabilized by <scene name='DOPA_decarboxylase/Vanderwaals/1'>van der waals contact</scene> with Ile-101 and Phe-103. The 4' hydroxyl group of the catechol ring participates in hydrogen bonding with <scene name='DOPA_decarboxylase/Thr-82/1'>Thr-82</scene>, further stabilizing the inhibitor in the active site cleft. PLP is further involved in substrate binding by forming a hydrogen bond to the 3' of the catechol ring. <scene name='DOPA_decarboxylase/His192/1'>His-192</scene>, a highly conserved residue of PLP-dependent decarboxylases <ref name="ishii">PMID:8889823 </ref> hydrogen bonds to the carboxylate group of carbiDOPA. |
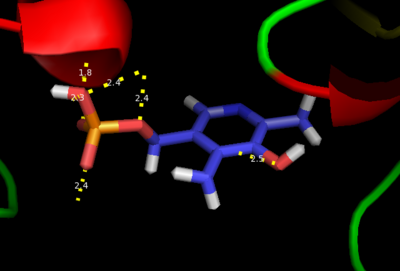
| + | [[image:actsite.png|thumb|center|400px|'''Key interactions between the active site residues, PLP, and carbiDOPA''']] | ||
---- | ---- | ||
| - | [[image:carbiDOPA.png|thumb|200px|'''carbiDOPA''']]The inhibitor <scene name='DOPA_decarboxylase/Carbidopa/1'>carbiDOPA</scene> binds to the enzyme by forming a hydrazone linkage with PLP through its hydrazine moiety. The catechol ring of carbiDOPA is deeply buried in the active site cleft and is stabilized by <scene name='DOPA_decarboxylase/Vanderwaals/1'>van der waals contact</scene> with Ile-101 and Phe-103. The 4' hydroxyl group of the catechol ring participates in hydrogen bonding with <scene name='DOPA_decarboxylase/Thr-82/1'>Thr-82</scene>, further stabilizing the inhibitor in the active site cleft. PLP is further involved in substrate binding by forming a hydrogen bond to the 3' of the catechol ring. <scene name='DOPA_decarboxylase/His192/1'>His-192</scene>, a highly conserved residue of PLP-dependent decarboxylases <ref name="ishii">PMID:8889823 </ref> hydrogen bonds to the carboxylate group of carbiDOPA. | ||
===Flexible Loop=== | ===Flexible Loop=== | ||
In all three crystal structures of DOPA decarboxylase solved to date, residues 328-339 are invisible in the electron density map. This is because these amino acids form a short mobile loop that is believed to be important to the catalytic mechanism of the enzyme <ref name="ishii">PMID:10082378 </ref>. During catalysis, this loop is proposed to lose its flexibility and extend toward the active site, both occluding the active site from solvent during catalysis and possibly even taking part in the catalytic mechanism. The mobile loop is found in other PLP-depended enzymes, such as glutamate 1-semialdehyde aminotransferase. The idea that this flexible loop plays an important role in catalysis is supported by the fact that it contains several highly conserved residues, Tyr-332 and Lys-334. The conformational change that occurs is thought to be a result of loop residues directly interacting with the inhbitor. | In all three crystal structures of DOPA decarboxylase solved to date, residues 328-339 are invisible in the electron density map. This is because these amino acids form a short mobile loop that is believed to be important to the catalytic mechanism of the enzyme <ref name="ishii">PMID:10082378 </ref>. During catalysis, this loop is proposed to lose its flexibility and extend toward the active site, both occluding the active site from solvent during catalysis and possibly even taking part in the catalytic mechanism. The mobile loop is found in other PLP-depended enzymes, such as glutamate 1-semialdehyde aminotransferase. The idea that this flexible loop plays an important role in catalysis is supported by the fact that it contains several highly conserved residues, Tyr-332 and Lys-334. The conformational change that occurs is thought to be a result of loop residues directly interacting with the inhbitor. | ||
Revision as of 01:42, 20 January 2012
| |||||||||||
3D structures of DOPA decarboxylase
Update November 2011
3k40 – DDC – Drosophila melanogaster
1js3 – pDDC + inhibitor – pig
1js6 - pDDC
3rbf, 3rbl – hDDC – human
3rch – hDDC + vitamin B6 phosphate + pyridoxal phosphate
References
- ↑ 1.0 1.1 Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000 Jan 15;8(1):R1-6. PMID:10673430
- ↑ Miles EW. The tryptophan synthase alpha 2 beta 2 complex. Cleavage of a flexible loop in the alpha subunit alters allosteric properties. J Biol Chem. 1991 Jun 15;266(17):10715-8. PMID:1904055
- ↑ Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001 Nov;8(11):963-7. PMID:11685243 doi:http://dx.doi.org/10.1038/nsb1101-963
- ↑ Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003 Sep;4(9):850-4. PMID:12949584 doi:http://dx.doi.org/10.1038/sj.embor.embor914
- ↑ Aurora R, Rose GD. Helix capping. Protein Sci. 1998 Jan;7(1):21-38. PMID:9514257 doi:10.1002/pro.5560070103
- ↑ Jansonius JN. Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol. 1998 Dec;8(6):759-69. PMID:9914259
- ↑ 7.0 7.1 Ishii S, Mizuguchi H, Nishino J, Hayashi H, Kagamiyama H. Functionally important residues of aromatic L-amino acid decarboxylase probed by sequence alignment and site-directed mutagenesis. J Biochem. 1996 Aug;120(2):369-76. PMID:8889823
Proteopedia Page Contributors and Editors (what is this?)
Brittany Todd, Michal Harel, David Canner, Alexander Berchansky, Brian Hernandez