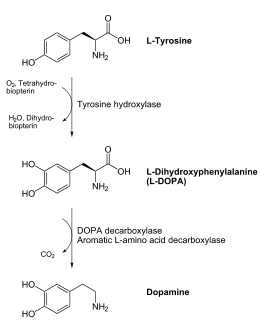
DOPA decarboxylase
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
---- | ---- | ||
===Overview=== | ===Overview=== | ||
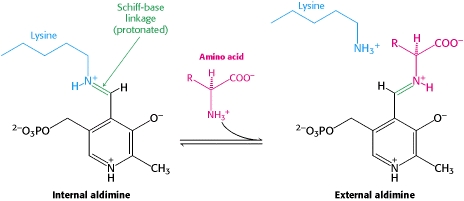
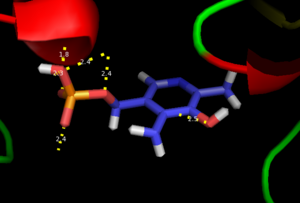
| - | Pyridoxal-5'-phosphate (PLP) is the biologically active phosphorylated derivative of vitamin B6. PLP-dependent enzymes are functionally diverse, with 150+ distinct activities, including transiminations, '''decarboxylations''', racemizations, and deaminations. With few exceptions, all PLP-dependent enzymes participate in amino acid metabolism. The versatility of PLP-dependent enzymes is, in large part, due to two basic chemical properties of the coenzyme. First, it can form a covalent bond with the substrate via its aldehyde group. Second, it can act as an electrophilic catalyst, serving as an electron sink which stabilizes the obligatory carbanionic intermediates formed during the reaction. In the resting state, PLP forms a covalent bond with the amino group of the active site Lysine (internal aldimine). Upon introduction of the substrate into the active site, a new Schiff base is generated (external aldimine). This transimination step is common to '''all''' PLP-dependent enzymes. | + | Pyridoxal-5'-phosphate (PLP) is the biologically active phosphorylated derivative of vitamin B6. PLP-dependent enzymes are functionally diverse, with 150+ distinct activities, including transiminations, '''decarboxylations''', racemizations, and deaminations. With few exceptions, all PLP-dependent enzymes participate in amino acid metabolism. The versatility of PLP-dependent enzymes is, in large part, due to two basic chemical properties of the coenzyme. First, it can form a covalent bond with the substrate via its aldehyde group. Second, it can act as an electrophilic catalyst, serving as an electron sink which stabilizes the obligatory carbanionic intermediates formed during the reaction. [[image:transimination.png|thumb|left|300px|'''transimination reaction''']]In the resting state, PLP forms a covalent bond with the amino group of the active site Lysine (internal aldimine). Upon introduction of the substrate into the active site, a new Schiff base is generated (external aldimine). This transimination step is common to '''all''' PLP-dependent enzymes. |
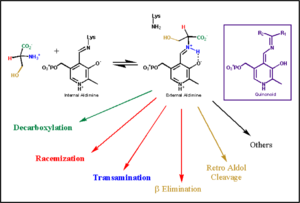
| - | Although these enzymes have wide range of function, | + | Although these enzymes have wide range of function, they can be classified into only five structural classes: the '''aspartate amino transferase family''', the tryptophan synthase β family, the alanine racemase family, the D-amino acid family, and the glycogen phosphorylase family. <ref name="percudani">PMID:12949584 </ref> <ref name="schneider">PMID:10673430 </ref> [[image:plp6.jpg|thumb|center|600px|'''The PLP is bound covalently to lysine residues in a Schiff base linkage (aldimine). In this form, it reacts with many free amino acids to replace the Schiff base to the lysine of the enzyme with a Schiff base to the amino acid substrate.'']] |
---- | ---- | ||
===The Aspartate Aminotransferase Family=== | ===The Aspartate Aminotransferase Family=== | ||
| Line 19: | Line 19: | ||
---- | ---- | ||
====Primary Structure==== | ====Primary Structure==== | ||
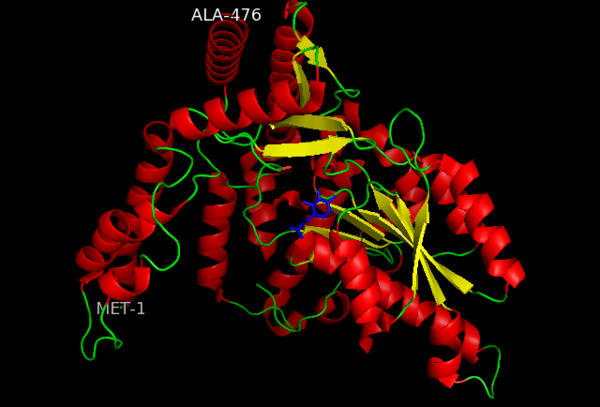
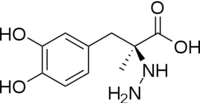
| - | The amino acid sequence of a proteins polypeptide chain is referred to as its primary structure. Each polypeptide chain of DOPA decarboxylase is composed of 486 amino acids that ultimately encode the three-dimensional structure of the protein. | + | The amino acid sequence of a proteins polypeptide chain is referred to as its primary structure. Each polypeptide chain of DOPA decarboxylase is composed of 486 amino acids that ultimately encode the three-dimensional structure of the protein. In 1991, the complete amino acid sequence was determined by peptide analysis, significantly increasing the structural and functional understanding of the enzyme <ref name="maras">PMID: 1935935 </ref>,. |
====Secondary Structure==== | ====Secondary Structure==== | ||
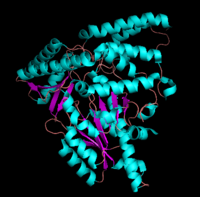
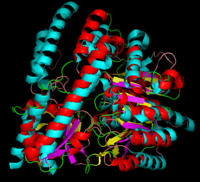
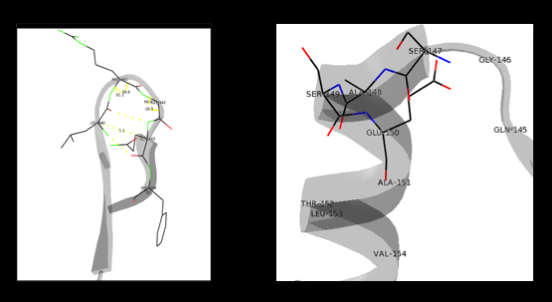
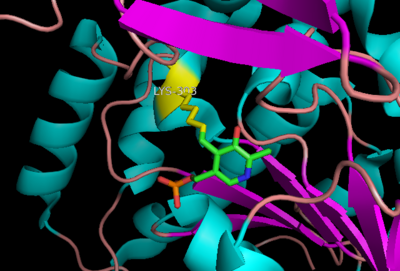
The formation of secondary structural elements (like α helices and β sheets) arise in response to the hydrophobic effect and the need to neutralize main-chain polar groups by '''hydrogen bonding'''. Each polypeptide chain of DOPA decarboxylase is composed of a seven-stranded mixed <scene name='DOPA_decarboxylase/Beta-sheet/1'>β sheet</scene>, a four-stranded anti-parallel <scene name='DOPA_decarboxylase/Beta-sheet2/1'>β sheet</scene>, several <scene name='DOPA_decarboxylase/Alpha_helices/1'>α helices</scene>, and other, lesser known, secondary structural elements (like loops and the extended strand). Another common secondary structure is the β-turn, or reverse turn. Depicted below is an example of a '''Type 1 β-turn''' of DOPA decarboxylase. This β-turn is comprised of residues Leu-440, Arg-441, Gly-442, and Gln-443. The distance between Cαi and Cαi+3 is 5.1Å, within the acceptable limit of 7Å. As in most β-turns, there is a hydrogen bond between the C=O of Leu-440 and the NH of Gln-443. The phi and psi angles of residues i+1 (Arg-441) and i+2 (Gly-442) are indicated in the diagram. | The formation of secondary structural elements (like α helices and β sheets) arise in response to the hydrophobic effect and the need to neutralize main-chain polar groups by '''hydrogen bonding'''. Each polypeptide chain of DOPA decarboxylase is composed of a seven-stranded mixed <scene name='DOPA_decarboxylase/Beta-sheet/1'>β sheet</scene>, a four-stranded anti-parallel <scene name='DOPA_decarboxylase/Beta-sheet2/1'>β sheet</scene>, several <scene name='DOPA_decarboxylase/Alpha_helices/1'>α helices</scene>, and other, lesser known, secondary structural elements (like loops and the extended strand). Another common secondary structure is the β-turn, or reverse turn. Depicted below is an example of a '''Type 1 β-turn''' of DOPA decarboxylase. This β-turn is comprised of residues Leu-440, Arg-441, Gly-442, and Gln-443. The distance between Cαi and Cαi+3 is 5.1Å, within the acceptable limit of 7Å. As in most β-turns, there is a hydrogen bond between the C=O of Leu-440 and the NH of Gln-443. The phi and psi angles of residues i+1 (Arg-441) and i+2 (Gly-442) are indicated in the diagram. | ||
Revision as of 15:50, 30 April 2012
| |||||||||||
3D structures of DOPA decarboxylase
Update November 2011
3k40 – DDC – Drosophila melanogaster
1js3 – pDDC + inhibitor – pig
1js6 - pDDC
3rbf, 3rbl – hDDC – human
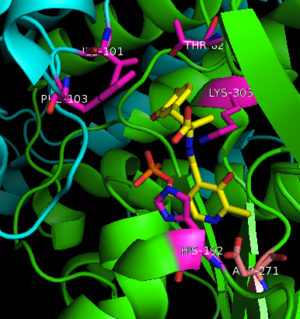
3rch – hDDC + vitamin B6 phosphate + pyridoxal phosphate
References
- ↑ 1.0 1.1 Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000 Jan 15;8(1):R1-6. PMID:10673430
- ↑ Miles EW. The tryptophan synthase alpha 2 beta 2 complex. Cleavage of a flexible loop in the alpha subunit alters allosteric properties. J Biol Chem. 1991 Jun 15;266(17):10715-8. PMID:1904055
- ↑ Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001 Nov;8(11):963-7. PMID:11685243 doi:http://dx.doi.org/10.1038/nsb1101-963
- ↑ Miles EW. The tryptophan synthase alpha 2 beta 2 complex. Cleavage of a flexible loop in the alpha subunit alters allosteric properties. J Biol Chem. 1991 Jun 15;266(17):10715-8. PMID:1904055
- ↑ Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003 Sep;4(9):850-4. PMID:12949584 doi:http://dx.doi.org/10.1038/sj.embor.embor914
- ↑ Maras B, Dominici P, Barra D, Bossa F, Voltattorni CB. Pig kidney 3,4-dihydroxyphenylalanine (dopa) decarboxylase. Primary structure and relationships to other amino acid decarboxylases. Eur J Biochem. 1991 Oct 15;201(2):385-91. PMID:1935935
- ↑ Aurora R, Rose GD. Helix capping. Protein Sci. 1998 Jan;7(1):21-38. PMID:9514257 doi:10.1002/pro.5560070103
- ↑ Jansonius JN. Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol. 1998 Dec;8(6):759-69. PMID:9914259
- ↑ 9.0 9.1 Ishii S, Mizuguchi H, Nishino J, Hayashi H, Kagamiyama H. Functionally important residues of aromatic L-amino acid decarboxylase probed by sequence alignment and site-directed mutagenesis. J Biochem. 1996 Aug;120(2):369-76. PMID:8889823
Proteopedia Page Contributors and Editors (what is this?)
Brittany Todd, Michal Harel, David Canner, Alexander Berchansky, Brian Hernandez