DOPA decarboxylase
From Proteopedia
(Difference between revisions)
| Line 53: | Line 53: | ||
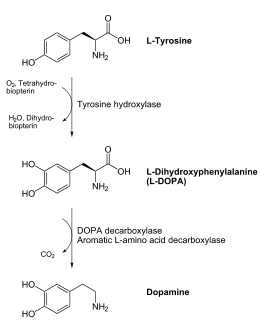
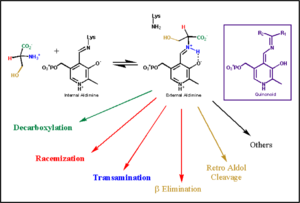
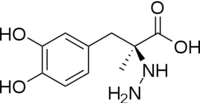
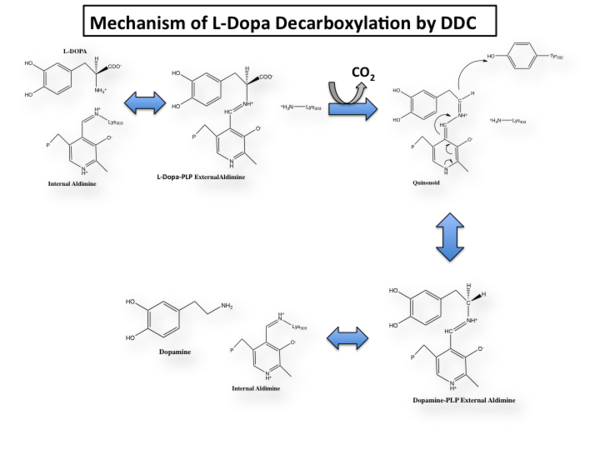
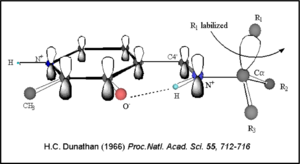
The mechanism of DDC catalyzed decarboxylation of L-Dopa to Dopamine has been well-studied due to the enzymes role in PD. Shown below is the mechanism of decarboxylation, as determined by several experimental approaches, including site-directed mutagenesis, UV-Vis Spectroscopy, X-Ray Crystallography, Multiple Sequence Alignment, and Stopped-Flow Spectroscopy. [[image:slide1.png|thumb|center|600px|]] Once again, the transimination step (conversion of internal to external aldimine) is common to all PLP-dependent enzymes. The unique absorption wavelengths of the PLP intermediates has allowed for their straightforward detection (for example, an internal Schiff base in the enolimine form absorbs at 310-330 nm, whereas the quinonoid intermediate absorbs at ~500 nm). The orientation of the quinonoid intermediate allows for stereospecific decarboxylation of the substrate at the alpha carbon, as predicted by '''Dunathan's stereoelectronic hypothesis''' <ref name="dunathan">PMID:224217 </ref>, in which he proposed that the substrate binds PLP such that the external aldimine intermediate is oriented perpendicular to the coenzyme pi bonding system. In doing so, the sigma-pi orbital overlap in the transition state is maximized, thus maximizing the rate of the reaction. | The mechanism of DDC catalyzed decarboxylation of L-Dopa to Dopamine has been well-studied due to the enzymes role in PD. Shown below is the mechanism of decarboxylation, as determined by several experimental approaches, including site-directed mutagenesis, UV-Vis Spectroscopy, X-Ray Crystallography, Multiple Sequence Alignment, and Stopped-Flow Spectroscopy. [[image:slide1.png|thumb|center|600px|]] Once again, the transimination step (conversion of internal to external aldimine) is common to all PLP-dependent enzymes. The unique absorption wavelengths of the PLP intermediates has allowed for their straightforward detection (for example, an internal Schiff base in the enolimine form absorbs at 310-330 nm, whereas the quinonoid intermediate absorbs at ~500 nm). The orientation of the quinonoid intermediate allows for stereospecific decarboxylation of the substrate at the alpha carbon, as predicted by '''Dunathan's stereoelectronic hypothesis''' <ref name="dunathan">PMID:224217 </ref>, in which he proposed that the substrate binds PLP such that the external aldimine intermediate is oriented perpendicular to the coenzyme pi bonding system. In doing so, the sigma-pi orbital overlap in the transition state is maximized, thus maximizing the rate of the reaction. | ||
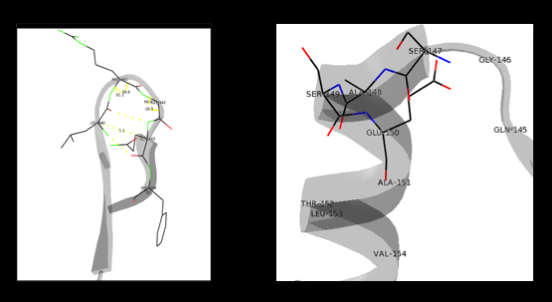
[[image:Dunathan.png|thumb|left|300px|'''Dunathan's Stereoeletronic Hypothesis, 1966''']] This way, the developing p orbital is aligned for maximal overlap with the extended p system, lowering the energy of the transition state and increasing the rate of the reaction. As well, by controlling substrate orientation, the enzyme can distinguish between '''deprotonation''' and '''decarboxylation'''. | [[image:Dunathan.png|thumb|left|300px|'''Dunathan's Stereoeletronic Hypothesis, 1966''']] This way, the developing p orbital is aligned for maximal overlap with the extended p system, lowering the energy of the transition state and increasing the rate of the reaction. As well, by controlling substrate orientation, the enzyme can distinguish between '''deprotonation''' and '''decarboxylation'''. | ||
| - | [[image:untitled.png|thumb|center|400px|]] | ||
===Mechanism Breakdown=== | ===Mechanism Breakdown=== | ||
| Line 59: | Line 58: | ||
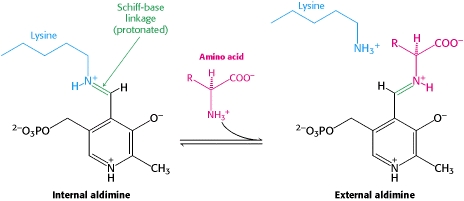
The first step of the reaction involves the binding of PLP to the enzyme via a Schiff base linkage between the aldehyde group of PLP and the ε—amino group of Lys303. An extensive hydrogen bond network further anchors PLP to the enzyme. | The first step of the reaction involves the binding of PLP to the enzyme via a Schiff base linkage between the aldehyde group of PLP and the ε—amino group of Lys303. An extensive hydrogen bond network further anchors PLP to the enzyme. | ||
====Step 2: Formation of an External Aldimine==== | ====Step 2: Formation of an External Aldimine==== | ||
| - | The second step of the reaction involves the binding of PLP with the substrate via a Schiff base linkage. The imine formed in the first step of the reaction is more succeptible than a free aldehyde to nucleophilic attack by the amino group of the substrate. Thus, not only does the internal aldimine between PLP and the Lys303 bind cofactor, it also serves to facilitate chemistry occurring in this second step. | + | The second step of the reaction involves the binding of PLP with the substrate via a Schiff base linkage. The imine formed in the first step of the reaction is more succeptible than a free aldehyde to nucleophilic attack by the amino group of the substrate. Thus, not only does the internal aldimine between PLP and the Lys303 bind cofactor, it also serves to facilitate chemistry occurring in this second step.[[image:untitled.png|thumb|center|400px|]] |
====Step 3: Formation of a Quinonoid Intermediate==== | ====Step 3: Formation of a Quinonoid Intermediate==== | ||
The formation of the quinonoid intermediate is common to all PLP-dependent enzymes, yet the orientation of the intermediate, as determined by key residues of the enzyme active site, determines the subsequent reaction (for example whether it will be a decarboxylation or transamination). A salt bridge that exists between Asp271 and the protonated pyridine nitrogen of PLP further enhances the ability of PLP to act as an electron sink and promote catalysis. As well, During the formation of the quinonoid intermediate, carbon dioxide is released. | The formation of the quinonoid intermediate is common to all PLP-dependent enzymes, yet the orientation of the intermediate, as determined by key residues of the enzyme active site, determines the subsequent reaction (for example whether it will be a decarboxylation or transamination). A salt bridge that exists between Asp271 and the protonated pyridine nitrogen of PLP further enhances the ability of PLP to act as an electron sink and promote catalysis. As well, During the formation of the quinonoid intermediate, carbon dioxide is released. | ||
| - | ====Step 4: Formation of an External Aldimine=== | + | ====Step 4: Formation of an External Aldimine==== |
The formation of the external aldimine between the product and PLP is the fourth step of the reaction. Here, Tyr332, with the assistance of His192, likely donates a proton to the quinonoid Cα intermediate. | The formation of the external aldimine between the product and PLP is the fourth step of the reaction. Here, Tyr332, with the assistance of His192, likely donates a proton to the quinonoid Cα intermediate. | ||
====Step 5: Formation of an Internal Aldimine and Product Release==== | ====Step 5: Formation of an Internal Aldimine and Product Release==== | ||
Revision as of 16:09, 1 May 2012
| |||||||||||
3D structures of DOPA decarboxylase
Update November 2011
3k40 – DDC – Drosophila melanogaster
1js3 – pDDC + inhibitor – pig
1js6 - pDDC
3rbf, 3rbl – hDDC – human
3rch – hDDC + vitamin B6 phosphate + pyridoxal phosphate
References
- ↑ 1.0 1.1 Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000 Jan 15;8(1):R1-6. PMID:10673430
- ↑ Miles EW. The tryptophan synthase alpha 2 beta 2 complex. Cleavage of a flexible loop in the alpha subunit alters allosteric properties. J Biol Chem. 1991 Jun 15;266(17):10715-8. PMID:1904055
- ↑ Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001 Nov;8(11):963-7. PMID:11685243 doi:http://dx.doi.org/10.1038/nsb1101-963
- ↑ Miles EW. The tryptophan synthase alpha 2 beta 2 complex. Cleavage of a flexible loop in the alpha subunit alters allosteric properties. J Biol Chem. 1991 Jun 15;266(17):10715-8. PMID:1904055
- ↑ Percudani R, Peracchi A. A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 2003 Sep;4(9):850-4. PMID:12949584 doi:http://dx.doi.org/10.1038/sj.embor.embor914
- ↑ Maras B, Dominici P, Barra D, Bossa F, Voltattorni CB. Pig kidney 3,4-dihydroxyphenylalanine (dopa) decarboxylase. Primary structure and relationships to other amino acid decarboxylases. Eur J Biochem. 1991 Oct 15;201(2):385-91. PMID:1935935
- ↑ Aurora R, Rose GD. Helix capping. Protein Sci. 1998 Jan;7(1):21-38. PMID:9514257 doi:10.1002/pro.5560070103
- ↑ Jansonius JN. Structure, evolution and action of vitamin B6-dependent enzymes. Curr Opin Struct Biol. 1998 Dec;8(6):759-69. PMID:9914259
- ↑ 9.0 9.1 Ishii S, Mizuguchi H, Nishino J, Hayashi H, Kagamiyama H. Functionally important residues of aromatic L-amino acid decarboxylase probed by sequence alignment and site-directed mutagenesis. J Biochem. 1996 Aug;120(2):369-76. PMID:8889823
- ↑ Hiscott JB, Defendi V. Simian virus 40 gene A regulation of cellular DNA synthesis. I. In permissive cells. J Virol. 1979 May;30(2):590-9. PMID:224217
Proteopedia Page Contributors and Editors (what is this?)
Brittany Todd, Michal Harel, David Canner, Alexander Berchansky, Brian Hernandez