User:Kristen Russo/Sandbox2 1N25
From Proteopedia
| Line 3: | Line 3: | ||
==Introduction== | ==Introduction== | ||
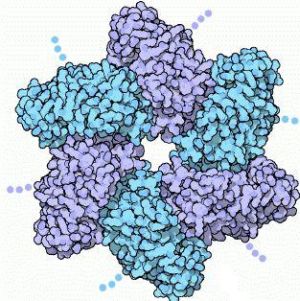
| - | Large T-antigen (LTag) is a viral [http://www.merriam-webster.com/medical/oncoprotein oncoprotein] derived from the polyomavirus SV40 (Simian virus 40). SV40 was first identified in the cultures of Asian rhesus monkeys in 1960. SV40 infects primate cells by forcing its way into the cells and releasing its DNA. Once inside, the DNA replicates and packages it in new viral capsids. SV40 uses the T-antigen protein to control both of these processes. There are three functional parts to the T-antigen. The first part is the LTag , which forms a six-fold ring by assembling with several other copies of the protein. The hole in the middle encircles a DNA double helix. | + | Large T-antigen (LTag) is a viral [http://www.merriam-webster.com/medical/oncoprotein oncoprotein] derived from the [http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.1000363 polyomavirus] SV40 (Simian virus 40). SV40 was first identified in the cultures of Asian rhesus monkeys in 1960. SV40 infects primate cells by forcing its way into the cells and releasing its DNA. Once inside, the DNA replicates and packages it in new viral capsids. SV40 uses the T-antigen protein to control both of these processes. There are three functional parts to the T-antigen. The first part is the LTag , which forms a six-fold ring by assembling with several other copies of the protein. The hole in the middle encircles a DNA double helix. |
[[Image:ltag.jpg|LTag]] | [[Image:ltag.jpg|LTag]] | ||
| Line 11: | Line 11: | ||
==How Does LTag's Structure Affect its Function?== | ==How Does LTag's Structure Affect its Function?== | ||
===Basic Structure=== | ===Basic Structure=== | ||
| - | <Structure load='1n25' size='500' frame='true' align='right' caption=' | + | <Structure load='1n25' size='500' frame='true' align='right' caption='LTag 1N25' scene='Insert optional scene name here' /> |
| - | LTag carries on several different functions that allow for viral replication. LTag [http://users.sdsc.edu/~youkha/duplication/_OLIGOMERS_RINGS/OLIGOMERIZATION/protein_oligomerization_how_why.pdf oligomerizes] into a double hexamer which binds to the viral origin of replication. This forms a prinitiation complex that melts, and ultimately unwinds the double-stranded DNA. After the formation of this complex, LTag works in coordination with certain host proteins (such as topoisomerase I) to promote bidirectional replication. The LTag monomner contains <scene name='User:Kristen_Russo/Sandbox2_1N25/Secondary_structure_alphabeta/1'>alpha helices and beta sheets</scene> that fold into three domains. The first domain <scene name='User:Kristen_Russo/Sandbox2_1N25/D1/5'>(D1)</scene> is the Zn domain that holds 5 alpha helices and is located at the N terminus. The second domain <scene name='User:Kristen_Russo/Sandbox2_1N25/D2/6'>(D2)</scene> folds into a core beta sheet that has five beta strands, sandwiched by alpha helices. The loop between alpha helix 9 and beta strand 1 is called the P-loop, and its function is to bind triphospahte for hydrolysis. The third domain (D3) contains seven alpha helices. Three of those form a circular ring at the <scene name='User:Kristen_Russo/Sandbox2_1N25/N_terminal/1'>N-terminal region</scene>, while the other four form a second circular ring at the <scene name='User:Kristen_Russo/Sandbox2_1N25/N_terminal_c_terminus/6'>C-terminus</scene>. The two rings are 'crosslinked' and are tightly packed against the P-loop. This forms a large globular bulge. The hexamer structure of LTag is formed from the three charged residues <scene name='User:Kristen_Russo/Sandbox2_1N25/Lys418_arg498_asp499/1'>LYS418, ARG498, and ASP499</scene>. These residues reside on one side of the molecule, and all point | + | LTag carries on several different functions that allow for viral replication. LTag [http://users.sdsc.edu/~youkha/duplication/_OLIGOMERS_RINGS/OLIGOMERIZATION/protein_oligomerization_how_why.pdf oligomerizes] into a double hexamer which binds to the viral origin of replication. This forms a prinitiation complex that melts, and ultimately unwinds the double-stranded DNA. After the formation of this complex, LTag works in coordination with certain host proteins (such as [http://www.callutheran.edu/BioDev/omm/topo1/topo1.htm topoisomerase I]) to promote bidirectional replication. The LTag monomner contains <scene name='User:Kristen_Russo/Sandbox2_1N25/Secondary_structure_alphabeta/1'>alpha helices and beta sheets</scene> that fold into three domains. The first domain <scene name='User:Kristen_Russo/Sandbox2_1N25/D1/5'>(D1)</scene> is the Zn domain that holds 5 alpha helices and is located at the N terminus. The second domain <scene name='User:Kristen_Russo/Sandbox2_1N25/D2/6'>(D2)</scene> folds into a core beta sheet that has five beta strands, sandwiched by alpha helices. The loop between alpha helix 9 and beta strand 1 is called the P-loop, and its function is to bind triphospahte for hydrolysis. The third domain (D3) contains seven alpha helices. Three of those form a circular ring at the <scene name='User:Kristen_Russo/Sandbox2_1N25/N_terminal/1'>N-terminal region</scene>, while the other four form a second circular ring at the <scene name='User:Kristen_Russo/Sandbox2_1N25/N_terminal_c_terminus/6'>C-terminus</scene>. The two rings are 'crosslinked', and are tightly packed against the P-loop. This forms a large globular bulge. The hexamer structure of LTag is formed from the three charged residues <scene name='User:Kristen_Russo/Sandbox2_1N25/Lys418_arg498_asp499/1'>LYS418, ARG498, and ASP499</scene>. These residues reside on one side of the molecule, and all point toward the P-loop. After the P-loop binds to ATP/ADP and Mg2+, the residues can bind to either the ATP/ADP or Mg+ ion, strengthening the interactions of the monomer thus forming a hexamer structure. <scene name='User:Kristen_Russo/Sandbox2_1N25/Arg498/1'>ARG498</scene> specifically, may have a role in ATP hydrolysis. |
===LTag Inhibits p53=== | ===LTag Inhibits p53=== | ||
| - | LTag forms a complex with p53 using its helicase domain (residues 251-677). The complex is composed of one LTag hexamer, binding with one monomeric p53 (which is 6 p53s). LTag <scene name='User:Kristen_Russo/Sandbox2_1N25/Helix_15/3'>alpha helix h15</scene> binds to the p53 region 1. This makes one assymetric unit, which | + | LTag forms a complex with p53 using its helicase domain (residues 251-677). The complex is composed of one LTag hexamer, binding with one monomeric p53 (which is 6 p53s). LTag <scene name='User:Kristen_Russo/Sandbox2_1N25/Helix_15/3'>alpha helix h15</scene> binds to the p53 region 1. This makes one assymetric unit, which is arranged in 2 copies. Considering that p53 region 1 may play a significant role when bound to DNA, it stabilizes the p53 tetramer, the binding of LTag may actually interefere with the [http://www.nature.com/onc/journal/v20/n21/abs/1204373a.html tetramerization]of p53. LTag's <scene name='User:Kristen_Russo/Sandbox2_1N25/H16/1'>h16</scene> residues binds to region 2 of p53 forming both [http://superhydrophobiccoating.com/hydrophilic-hydrophobic-8/ hydrophilic and hydrophobic] side chains. The most significant interaction in region 2 is LTag's <scene name='User:Kristen_Russo/Sandbox2_1N25/Asp402/3'>ASP402</scene> binding to and formination of a salt bridge with p53's ARG248. In human cancers, this ARG248 is known to be the most frequently mutated residue, which leads to the assumption that the binding of LTag could be the reason for this mutation. Within this region, LTag also has a hydrophobic <scene name='User:Kristen_Russo/Sandbox2_1N25/Pocket/1'>pocket</scene>, which is composed of residues LEU609, TYR612, TRP581, and TYR582. This pocket forms a strong hydrophobic interaction to the p53 residue MET246, and it plays a very important role in the overall binding of LTag to p53. In addition to the binding within the pocket, <scene name='User:Kristen_Russo/Sandbox2_1N25/Pro584/2'>PRO584</scene> also packs with the main chain of p53 around its residue MET243. |
==Why is it Important?== | ==Why is it Important?== | ||
Revision as of 16:05, 8 May 2012
Contents |
Large T-Antigen
|
Introduction
Large T-antigen (LTag) is a viral oncoprotein derived from the polyomavirus SV40 (Simian virus 40). SV40 was first identified in the cultures of Asian rhesus monkeys in 1960. SV40 infects primate cells by forcing its way into the cells and releasing its DNA. Once inside, the DNA replicates and packages it in new viral capsids. SV40 uses the T-antigen protein to control both of these processes. There are three functional parts to the T-antigen. The first part is the LTag , which forms a six-fold ring by assembling with several other copies of the protein. The hole in the middle encircles a DNA double helix.
The other two functional parts of the T-antigen are: the central domain which has a small patch and anchors the T-antigen to the SV40 genome by binding to the regulatory region, and the third domain which interacts with cellular proteins, therefore directing the different stages of the viral life cycle. There are 12 copies of the T-antigen protein which forms a long tube around the DNA. Although there are three parts to the T-antigen, LTag is the fragment that seems to be associated with the formation of cancer. LTag is a member of the helicase family III, and has been known to hinder the function of cellular tumor suppressors, such as p53. P53 is important because it has the ability to either stop the cell cycle and allow for DNA repair, or to cause apoptosis. This p53 gene is found mutated in more than 50% of human cancers. LTag targets and inhibits p53 function, which ends up deregulating cellular growth control.
How Does LTag's Structure Affect its Function?
Basic Structure
|
LTag carries on several different functions that allow for viral replication. LTag oligomerizes into a double hexamer which binds to the viral origin of replication. This forms a prinitiation complex that melts, and ultimately unwinds the double-stranded DNA. After the formation of this complex, LTag works in coordination with certain host proteins (such as topoisomerase I) to promote bidirectional replication. The LTag monomner contains that fold into three domains. The first domain is the Zn domain that holds 5 alpha helices and is located at the N terminus. The second domain folds into a core beta sheet that has five beta strands, sandwiched by alpha helices. The loop between alpha helix 9 and beta strand 1 is called the P-loop, and its function is to bind triphospahte for hydrolysis. The third domain (D3) contains seven alpha helices. Three of those form a circular ring at the , while the other four form a second circular ring at the . The two rings are 'crosslinked', and are tightly packed against the P-loop. This forms a large globular bulge. The hexamer structure of LTag is formed from the three charged residues . These residues reside on one side of the molecule, and all point toward the P-loop. After the P-loop binds to ATP/ADP and Mg2+, the residues can bind to either the ATP/ADP or Mg+ ion, strengthening the interactions of the monomer thus forming a hexamer structure. specifically, may have a role in ATP hydrolysis.
LTag Inhibits p53
LTag forms a complex with p53 using its helicase domain (residues 251-677). The complex is composed of one LTag hexamer, binding with one monomeric p53 (which is 6 p53s). LTag binds to the p53 region 1. This makes one assymetric unit, which is arranged in 2 copies. Considering that p53 region 1 may play a significant role when bound to DNA, it stabilizes the p53 tetramer, the binding of LTag may actually interefere with the tetramerizationof p53. LTag's residues binds to region 2 of p53 forming both hydrophilic and hydrophobic side chains. The most significant interaction in region 2 is LTag's binding to and formination of a salt bridge with p53's ARG248. In human cancers, this ARG248 is known to be the most frequently mutated residue, which leads to the assumption that the binding of LTag could be the reason for this mutation. Within this region, LTag also has a hydrophobic , which is composed of residues LEU609, TYR612, TRP581, and TYR582. This pocket forms a strong hydrophobic interaction to the p53 residue MET246, and it plays a very important role in the overall binding of LTag to p53. In addition to the binding within the pocket, also packs with the main chain of p53 around its residue MET243.
Why is it Important?
Medical Relevance
Although SV40 originated in monekys, it is believed that the inoculation of about 98 million people back in 1960 with a contaminated polio vaccine was the major route of SV40 into humans. Today, SV40 DNA sequences are found in 50-70% of mesotheliomas and 70-90% of ependymomas. Many experiments are done to see if SV40 is present in human peripheral blood samples, and also whether it plays a role in human leukemias and lymphomas. In a study where 266 samples of human blood samples were studied, SV40 was found in 7 of 11 patients with AIDS related lymphomas. Not only was SV40 found, but the T-antigen signal was clear while the VP (a different region of the SV40 protein) was insignificant.
Conclusion
References
Arrington, A. S., Moore, M. S., & Butel, J. S. (2003). Sv40-positive brain tumor in scientist with risk of laboratory exposure to the virus. Short Report, 23, 2231-2235.
Cuesta, I., Nunez-Ramirez, R., Scheres, S. H. W., Gai, D., Chen, X. S., Fanning, E., & Carazo, J. M. (2010). Conformational rearrangements of sv40 large t antigen during early replication events. Science Direct, 397, 1276-1286.
David, H., Mendoza, S., Konishi, T., & Miller, C. W. (2000). Simian virus 40 is present in human lymphomas and normal blood. Cancer Letters, 57-64.
Li, D., Zhao, R., Lilyestrom, W., Gai, D., Zhang, R., DeCaprio, J. A., Fanning, E., Jochimiak, A., Szakonyi, G., & Chen, X.S., (2003). Structure of the replicative helicase of the oncoprotein sv40 large tumour antigen. Nature, 423(6939), 512-518.
Lilyestrom, W., Klein, M. G., Zhang, R., Joachimiak, A., & Chen, X. S. (2006). Crystal structure of sv40 large t-antigen bound to p53: Interplay between a viral oncoprotein and a cellular tumor suppressor. Genes and Development, 2373-2382.
Pepper, C., Jasani, B., Navabi, H., Wynford-Thomas, D., & Gibbs, A. R. (1996). Simian virus 40 large t antigen primer specific dna amplification in human pleural mesothelioma tissue. Rapid Communication, 51, 1074-1076.