This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
1d66
From Proteopedia
m (Protected "1d66" [edit=sysop:move=sysop]) |
|||
| Line 1: | Line 1: | ||
[[Image:1d66.png|left|200px]] | [[Image:1d66.png|left|200px]] | ||
| - | <!-- | ||
| - | The line below this paragraph, containing "STRUCTURE_1d66", creates the "Structure Box" on the page. | ||
| - | You may change the PDB parameter (which sets the PDB file loaded into the applet) | ||
| - | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | ||
| - | or leave the SCENE parameter empty for the default display. | ||
| - | --> | ||
{{STRUCTURE_1d66| PDB=1d66 | SCENE= }} | {{STRUCTURE_1d66| PDB=1d66 | SCENE= }} | ||
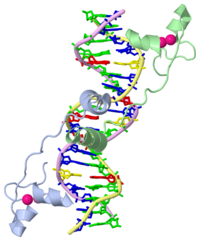
===DNA RECOGNITION BY GAL4: STRUCTURE OF A PROTEIN/DNA COMPLEX=== | ===DNA RECOGNITION BY GAL4: STRUCTURE OF A PROTEIN/DNA COMPLEX=== | ||
| - | |||
| - | <!-- | ||
| - | The line below this paragraph, {{ABSTRACT_PUBMED_1557122}}, adds the Publication Abstract to the page | ||
| - | (as it appears on PubMed at http://www.pubmed.gov), where 1557122 is the PubMed ID number. | ||
| - | --> | ||
{{ABSTRACT_PUBMED_1557122}} | {{ABSTRACT_PUBMED_1557122}} | ||
| Line 23: | Line 12: | ||
==See Also== | ==See Also== | ||
*[[Gal4|Gal4]] | *[[Gal4|Gal4]] | ||
| - | *[[Gal4 (Hebrew)|Gal4 (Hebrew)]] | ||
| - | *[[Getting Unremediated PDB Files|Getting Unremediated PDB Files]] | ||
*[[Hydrogen in macromolecular models|Hydrogen in macromolecular models]] | *[[Hydrogen in macromolecular models|Hydrogen in macromolecular models]] | ||
| - | *[[Jmol/Application|Jmol/Application]] | ||
| - | *[[Molecular Playground/Procedures|Molecular Playground/Procedures]] | ||
| - | *[[User:Amir Mitchell/hebrew Gal4|User:Amir Mitchell/hebrew Gal4]] | ||
| - | *[[User:Amir Mitchell/hebrew IPNS|User:Amir Mitchell/hebrew IPNS]] | ||
*[[User:Wayne Decatur/Biochem642 Molecular Visualization 2010 Fall Sessions|User:Wayne Decatur/Biochem642 Molecular Visualization 2010 Fall Sessions]] | *[[User:Wayne Decatur/Biochem642 Molecular Visualization 2010 Fall Sessions|User:Wayne Decatur/Biochem642 Molecular Visualization 2010 Fall Sessions]] | ||
*[[User:Wayne Decatur/Biochem642 Molecular Visualization Sessions|User:Wayne Decatur/Biochem642 Molecular Visualization Sessions]] | *[[User:Wayne Decatur/Biochem642 Molecular Visualization Sessions|User:Wayne Decatur/Biochem642 Molecular Visualization Sessions]] | ||
Revision as of 11:15, 26 July 2012
| |||||||||
| 1d66, resolution 2.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
DNA RECOGNITION BY GAL4: STRUCTURE OF A PROTEIN/DNA COMPLEX
A specific DNA complex of the 65-residue, N-terminal fragment of the yeast transcriptional activator, GAL4, has been analysed at 2.7 A resolution by X-ray crystallography. The protein binds as a dimer to a symmetrical 17-base-pair sequence. A small, Zn(2+)-containing domain recognizes a conserved CCG triplet at each end of the site through direct contacts with the major groove. A short coiled-coil dimerization element imposes 2-fold symmetry. A segment of extended polypeptide chain links the metal-binding module to the dimerization element and specifies the length of the site. The relatively open structure of the complex would allow another protein to bind coordinately with GAL4.
DNA recognition by GAL4: structure of a protein-DNA complex., Marmorstein R, Carey M, Ptashne M, Harrison SC, Nature. 1992 Apr 2;356(6368):408-14. PMID:1557122
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
About this Structure
1d66 is a 4 chain structure of Gal4 and Hydrogen in macromolecular models with sequence from Saccharomyces cerevisiae. Full crystallographic information is available from OCA.
See Also
- Gal4
- Hydrogen in macromolecular models
- User:Wayne Decatur/Biochem642 Molecular Visualization 2010 Fall Sessions
- User:Wayne Decatur/Biochem642 Molecular Visualization Sessions
Reference
- Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408-14. PMID:1557122 doi:http://dx.doi.org/10.1038/356408a0