Sandbox SouthUniversity1
From Proteopedia
| Line 10: | Line 10: | ||
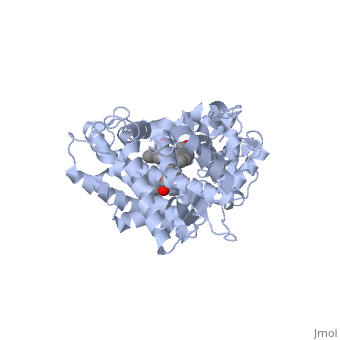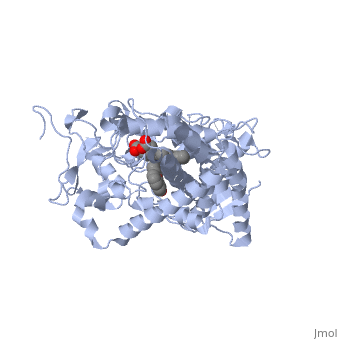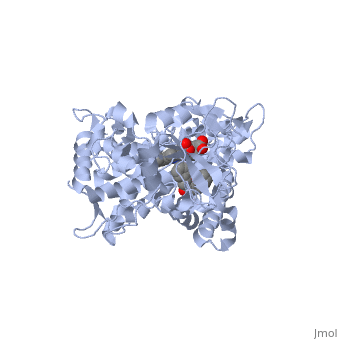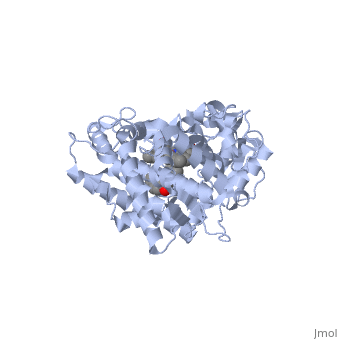
Now look at the substrate molecule being oxidized and view its orientation relative to the heme group by clicking <scene name='Sandbox_SouthUniversity1/Heme_and_flavone/3'>this link</scene> Resize and rotate the molecules until you can see how the two molecules are oriented in relationship to each other. The flavone is metabolized (oxidized) by introduction of a hydroxy group onto the phenyl ring that is attached to the tricyclic ring system. | Now look at the substrate molecule being oxidized and view its orientation relative to the heme group by clicking <scene name='Sandbox_SouthUniversity1/Heme_and_flavone/3'>this link</scene> Resize and rotate the molecules until you can see how the two molecules are oriented in relationship to each other. The flavone is metabolized (oxidized) by introduction of a hydroxy group onto the phenyl ring that is attached to the tricyclic ring system. | ||
| + | |||
| + | |||
| + | Purely based on the distance between the pheny ring of the flavone substrate and the heme iron, oxidation might be expected to take place at the 4 (para) position. However, other factors also help determine the regioselectivity of the metabolism (selectivity for oxidation at the different possible positions). One of these factors is the relative reactivity of the various positions on the substrate. | ||
| + | |||
| + | </StructureSection> | ||
<quiz display=simple> | <quiz display=simple> | ||
| Line 21: | Line 26: | ||
|| You would not expect this to be likely, based on the distance from the active iron center to either of the ortho positions. | || You would not expect this to be likely, based on the distance from the active iron center to either of the ortho positions. | ||
</quiz> | </quiz> | ||
| - | |||
| - | Purely based on the distance between the pheny ring of the flavone substrate and the heme iron, oxidation might be expected to take place at the 4 (para) position. However, other factors also help determine the regioselectivity of the metabolism (selectivity for oxidation at the different possible positions). One of these factors is the relative reactivity of the various positions on the substrate. | ||
| - | |||
| - | </StructureSection> | ||
| - | |||
| - | |||
| Line 58: | Line 57: | ||
| + | <quiz display=simple> | ||
| + | {Question | ||
| + | |type="()"} | ||
| + | + The correct answer. | ||
| + | || Feedback for correct answer. | ||
| + | - Distractor. | ||
| + | || Feedback for distractor. | ||
| + | - Distractor. | ||
| + | || Feedback for distractor. | ||
| + | - Distractor. | ||
| + | || Feedback for distractor. | ||
| + | </quiz> | ||
Revision as of 19:50, 5 June 2012
==Selectivity of Drug Metabolism by Cytochrome P450 Enzymes==
| |||||||||||
Different members of the CYP450 enzymes preferentially metabolize different xenobiotics. What features of a drug do you think might cause it to be metabolized by one CYP versus another? Draft: <illustration of residues in active site> Draft animation Draft animation
draft: <insert quiz here on electrostatic and steric factors>
draft: <insert animations here contrasting the shape of the active site of CYP3A4 and e.g. CYP2E1>
draft: <insert examples of substrates for 2 different CYPs, overlay them on top of each other illustrate the molecular commonalities>
draft <illustrate the size of the binding pockets and their electronic character>
draft test of scene names:
<Draft Quiz>
References The structure of CYP1A2 shown above is found in the Research Collaboration of Structural Bioinformatics Protein Database as entry 2HI4.