This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
1cma
From Proteopedia
| Line 1: | Line 1: | ||
| - | [[Image:1cma.jpg|left|200px]] | + | [[Image:1cma.jpg|left|200px]] |
| - | + | ||
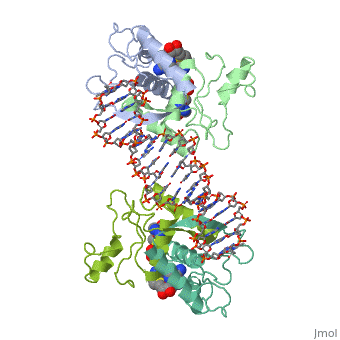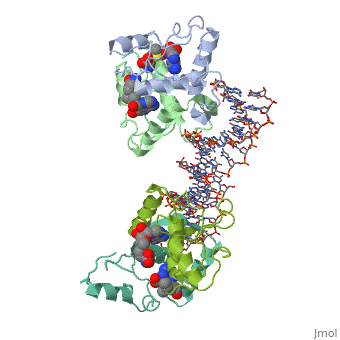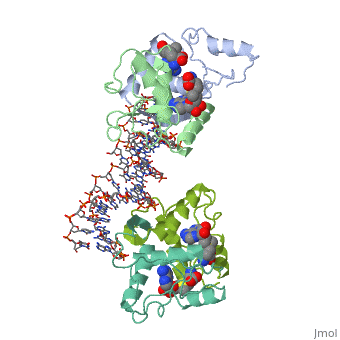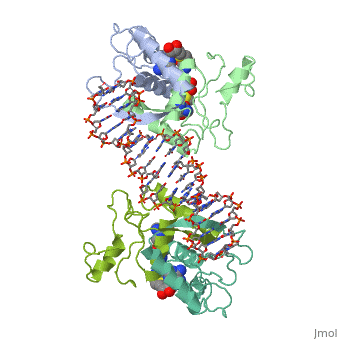
| - | '''MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE''' | + | {{Structure |
| + | |PDB= 1cma |SIZE=350|CAPTION= <scene name='initialview01'>1cma</scene>, resolution 2.800Å | ||
| + | |SITE= | ||
| + | |LIGAND= <scene name='pdbligand=SAM:S-ADENOSYLMETHIONINE'>SAM</scene> | ||
| + | |ACTIVITY= | ||
| + | |GENE= | ||
| + | }} | ||
| + | |||
| + | '''MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE''' | ||
| + | |||
==Overview== | ==Overview== | ||
| Line 7: | Line 16: | ||
==About this Structure== | ==About this Structure== | ||
| - | 1CMA is a [ | + | 1CMA is a [[Single protein]] structure of sequence from [http://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1CMA OCA]. |
==Reference== | ==Reference== | ||
| - | Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands., Somers WS, Phillips SE, Nature. 1992 Oct 1;359(6394):387-93. PMID:[http:// | + | Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands., Somers WS, Phillips SE, Nature. 1992 Oct 1;359(6394):387-93. PMID:[http://www.ncbi.nlm.nih.gov/pubmed/1406951 1406951] |
[[Category: Escherichia coli]] | [[Category: Escherichia coli]] | ||
[[Category: Single protein]] | [[Category: Single protein]] | ||
| Line 19: | Line 28: | ||
[[Category: protein-dna complex]] | [[Category: protein-dna complex]] | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Mar 20 10:26:09 2008'' |
Revision as of 08:26, 20 March 2008
| |||||||
| , resolution 2.800Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | |||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
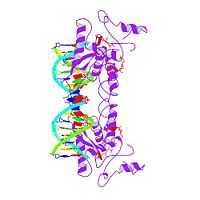
MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE
Overview
The crystal structure of the met repressor-operator complex shows two dimeric repressor molecules bound to adjacent sites 8 base pairs apart on an 18-base-pair DNA fragment. Sequence specificity is achieved by insertion of double-stranded antiparallel protein beta-ribbons into the major groove of B-form DNA, with direct hydrogen-bonding between amino-acid side chains and the base pairs. The repressor also recognizes sequence-dependent distortion or flexibility of the operator phosphate backbone, conferring specificity even for inaccessible base pairs.
About this Structure
1CMA is a Single protein structure of sequence from Escherichia coli. Full crystallographic information is available from OCA.
Reference
Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands., Somers WS, Phillips SE, Nature. 1992 Oct 1;359(6394):387-93. PMID:1406951
Page seeded by OCA on Thu Mar 20 10:26:09 2008